| |
Predicting SVR/Nonresponse at Weeks 1, 4, 12 with Pegasys/RBV
|
| |
| |
"Hepatitis C virus RNA kinetics during the initial 12 weeks treatment with pegylated interferon-alpha 2a (Pegasys) and ribavirin according to virological response"
T. Carlsson1, O. Reichard1, G. Norkrans2, J. Bläckberg3, P. Sangfelt4, E. Wallmark5 and O. Weiland1
1Division of Infectious Diseases, Karolinska University Hospital (Solna and Huddinge), Karolinska Institutet, Stockholm; 2Sahlgrenska University Hospital/East, Gothenburg University, Gothenburg; 3Lunds University Hospital, Lund University, Lund; 4Uppsala Akademiska Hospital, Uppsala University, Uppsala; and 5Malmö University Hospital, Lund University, Malmö, Sweden
During the last decade, considerable improvements have been made in the treatment of chronic hepatitis C. Sustained virologic response (SVR) rates have risen from 10-20% overall with interferon alpha (IFN) monotherapy [1,2] to 50-80%, depending on genotype, with pegylated IFN (PEG-IFN) plus ribavirin [3-5].
The daily production of hepatitis C virus (HCV) is estimated to be 1012 virions [6,7]. During IFN treatment HCV RNA levels decline in a biphasic manner [7]. The first phase noted in most patients is dependent on the IFN dose and lasts 24-48 h. The second phase varies from days to months in length, and is assumed to reflect elimination of infected hepatocytes [7]. During this phase, HCV RNA levels continue to decline in responders but not in nonresponders. The HCV RNA decline is usually more rapid in genotype non-1- than 1-infected patients [8-10] and steeper with higher IFN doses [11].
A >3-log10 decline within the first 4 weeks of treatment predicts a high likelihood of an SVR [12]. On the other hand if a 2-log10 drop in HCV RNA has not been reached by week 12 during therapy, the negative predictive value (NPV) for achieving an SVR is close to 100% [3,13].
Induction therapy as a strategy has been tried to achieve more rapid decrease in HCV RNA levels, and higher SVR rates, but so far with only limited success [10,14-16].
The aim of the present study was to follow the HCV RNA kinetics during the initial 12 weeks of treatment with PEG-IFN plus ribavirin, and to correlate the early decrease in HCV RNA with virological outcomes at the end of follow-up.
Summary. To optimize treatment of chronic hepatitis C early identification of patients who will not achieve a sustained virological response (SVR) is desirable. We investigated hepatitis C virus (HCV) RNA kinetics at day 1 (in 15 patients; genotypes 1 and non-1, 9 and 6 respectively) at weeks 1, 4 and 12 (in 53 patients; genotypes 1 and non-1, 19 and 34, respectively) during treatment with pegylated interferon alpha-2a and ribavirin. Patients with SVR had a significantly more pronounced mean log10 decline from baseline in HCV RNA levels at weeks 1 and 4 compared with patients who failed to achieve SVR (1.99 vs 0.85 at week 1, P = 0.0003 and 2.89 vs 1.72 at week 4, P = 0.0159), whereas no difference was noted after day 1.
For patients with a 2-log10 decrease in HCV RNA levels at day 7, the positive predictive value (PPV) for a SVR was 92%, whereas week 12 was the best time point for predicting a later nonresponse [negative predictive value (NPV) 92%] in patients failing to achieve a 2-log10 drop.
For patients with genotype non-1 and a 2-log10 decrease in HCV RNA levels the PPV for a SVR was 89% week 1, and 79% weeks 4 and 12.
The corresponding NPV for patients with genotype non-1 were 43, 40 and 100% respectively.
During treatment with pegylated interferon alpha-2a plus ribavirin the HCV RNA decline at week 1 was an accurate predictor of SVR in patients who had achieved a 2-log10 drop in HCV RNA levels, whereas the lack of such decline week 12 was an accurate marker of a nonresponse.
AUTHOR DISCUSSION
Treatment of chronic HCV infection has become more effective during recent years, but many patients still do not clear their infection. Such patients may experience considerable treatment-related side effects, and the treatment is expensive. Early treatment withdrawal is desirable for nonresponding patients. Treatment week 24 was initially considered to be the best time point for prediction of a nonresponse. Presently, however, treatment week 12 is considered to be the most appropriate time point. Thus, a less than 2-log10 drop in HCV RNA levels at this time point can predict a nonresponse with almost 100% certainty [3,13,23]. On the contrary, prediction of a nonresponse has been possible after only one dose of standard IFN in a subset of patients [10,24]. In the present study, HCV RNA levels did not differ significantly at day 1 between sustained virological responders and patients lacking such response in the subgroup analysed also day 1. Already day 7, however, before the second PEG IFN alpha-2a dose was given, a significant difference was detected (P = 0.02). This difference was also noted at week 4 (P = 0.0293; Fig. 2a), and when only genotype 1-infected patients with SVR and response/relapse were compared with nonresponders (P = 0.02 and P = 0.045, respectively).
Fig. 2 (a) Individual hepatitis C virus RNA levels in patients with genotype 1 infection at day 1, 7 and 28 of treatment with pegylated interferon a-2a and ribavirin. SR indicates sustained virologic responders; NR, nonresponders; RR, responder/relapsers. (b) Individual hepatitis C virus RNA levels in patients with genotype non-1 infection at day 1, 7 and 28 of treatment with pegylated interferon a-2a and ribavirin. SR indicates sustained virologic responders. (c) The median hepatitis C virus RNA level (bar within the box) in patients with genotype 1 and genotype non-1 infection at days 1, 7 and 28 of treatment with pegylated interferon a-2a and ribavirin (open boxes = genotype 1-infected: n = 9; grey boxes = genotype non-1-infected: n = 6). The boxes show the 25th to 75th percentiles, and the lower and upper bars show the 10th and 90th percentiles, respectively.
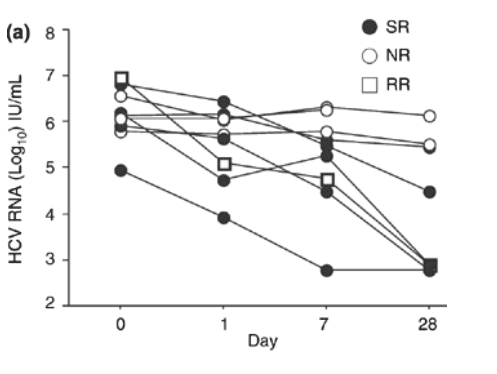
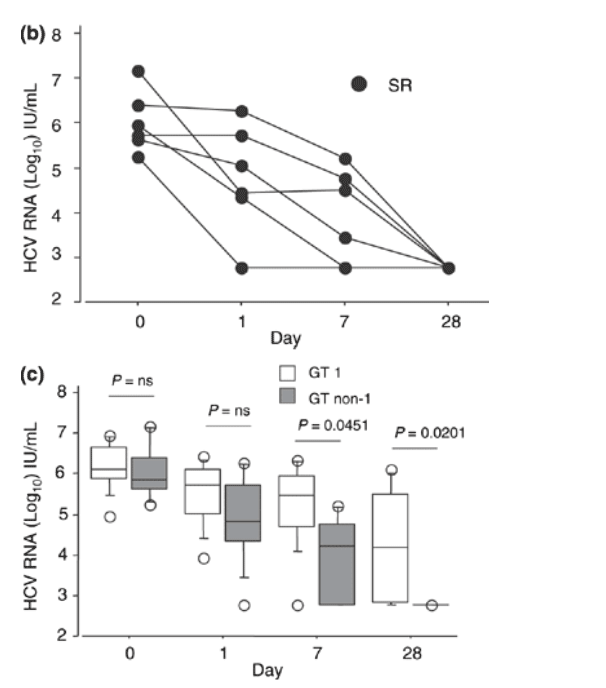
A difference in HCV RNA decline according to genotype was also noted. Hence, patients with genotype non-1 infection had a significantly more pronounced decline in HCV RNA levels at week 1 than patients with genotype 1 infection (P = 0.0292).
Prediction of final response was possible in the subset of patients who achieved a >2-log10 drop in HCV RNA levels at week 1 or 4 during treatment, when these patients had a PPV of 92 and 77%, respectively, for an SVR (Table 4).
Correspondingly, sustained virological responders among our 53 patients had a significantly more pronounced decline in HCV RNA levels at weeks 1 and 4 compared with nonsustained responders (P = 0.0001 and P = 0.0004, respectively), as noted earlier [10,13]. The same findings were also noted in genotype 1-infected patients when analysed separately. Among patients with genotype non-1 the majority achieved an SVR; therefore, no meaningful statistical comparison could be made between sustained virological responders and nonresponders in this group. Furthermore, as up to 80-90% of genotype 2- and 3-infected patients will clear their infection with the present treatment regimens [3-5], the most cost-effective therapy for these patients might be treatment for 24 weeks without monitoring the HCV RNA levels during treatment.
The HCV RNA levels decline in two phases during IFN treatment [9]. Some researchers have failed to notice this during PEG IFN treatment [25], whereas it was noted when PEG IFN was combined with ribavirin by others [26]. We could not detect a significant difference in the decline of HCV RNA levels between patients with SVR and patients lacking SVR day 1 after the first PEG IFN alpha-2a dose. PEG IFN has a slower absorption rate than standard IFN, which means that it takes a longer time for the agent to reach maximal serum concentration [27]. Because of this, we may have missed a difference in HCV RNA decline before day 7 which we might have detected if the HCV RNA levels had been analysed day 2 or 3. However, the HCV RNA decline day 7 after a single dose of PEG-IFN plus ribavirin offered a chance to predict a later SVR, with a PPV of 92% if a 2-log10 HCV RNA decline had been achieved, whereas a nonsustained response could not be predicted with a high NPV earlier than week 12 in accordance with what others have found [13,28].
In conclusion, we found that during treatment with PEG IFN alpha-2a plus ribavirin, analysis of HCV RNA levels day 1 was not useful as a predictor of treatment outcome, in contrast to findings during treatment with standard IFN and ribavirin. Week 1, on the contrary, can be used with greater accuracy for predicting an SVR in the subgroup of patients who have achieved a 2-log10 drop in HCV RNA levels, and week 12 for predicting a nonresponse if such a drop has not been reached.
RESULTS
Sixty patients were recruited. One patient discontinued treatment after the second dose of PEG IFN alpha-2a and was excluded from further analysis. Four patients discontinued treatment prematurely, one each at treatment weeks 8, 12 and 27 for unknown reasons, and one at week 16 for psychiatric reasons. These four patients plus one other patient, who was HCV RNA-negative after 48 weeks of treatment but was lost to follow-up, were excluded from the kinetic analysis. The only patient with genotype 4 infection was also excluded in order not to influence the comparison between genotype 1 and genotype non-1 (i.e. genotype 2 and 3) infections. One additional patient discontinued treatment at week 16 because of arthralgias but was negative for HCV RNA at follow-up and was considered to be a sustained virological responder in the analysis. Thus, the analysis of HCV RNA kinetics during the initial 12 weeks of treatment was based on 53 patients. In three of these patients, the PEG IFN alpha-2a dose was reduced because of thrombocytopenia or neutropenia, and in three others, the ribavirin dose because of anaemia. Baseline demographic data are given in Table 1. No difference in baseline characteristics were noted between genotype 1- (n = 19) and genotype non-1-infected patients (n = 34) except for baseline mean ALT levels, which were higher in genotype non-1-infected patients (P = 0.009).
Overall virologic outcome according to genotype--
The total study group included 53 patients, of whom 34 (64%) achieved a SVR (eight genotype 1; 26 genotype non-1), 19 (36%) a nonsustained response [response/relapse, n = 10 [3 genotype 1; 7 genotype non-1)] and nine (17%) a nonresponse (8 genotype 1; 1 genotype non-1).
Among genotype 1-infected patients, eight of 19 (42%) achieved an SVR vs 26 of 34 (76%) of the genotype non-1-infected patients (genotype 2 or 3; P = 0.0127). Among the 19 genotype 1-infected patients two of nine treated for 24 weeks compared with six of 10 treated for 48 weeks achieved an SVR (P = 0.1).
HCV RNA decline in individual patients--
The mean log10 HCV RNA decline in genotype 1-infected patients with SVR was greater at week 1 and week 4 (from 1.6 x 106 IU/mL at baseline to 0.14 x 106 IU/mL at week 1 and 0.0006 x 106 IU/mL at week 4, respectively) than in patients with nonresponse (from 1.35 x 106 to 1.5 x 106 and 0.84 x 106 IU/mL, respectively; baseline; P = NS, week 1; P = 0.0018 and week 4; P = 0.0026) (Fig. 1a). Responder/relapsers also had a greater HCV RNA decline at week 1 and week 4 (from 2.1 x 106 IU/mL at baseline to 0.085 x 106 IU/mL at week 1 and 0.0068 x 106 IU/mL at week 4, respectively) compared with nonresponders (from 1.35 x 106 to 1.5 x 106 and 0.84 x 106 IU/mL, respectively; baseline; P = NS, week 1; P = 0.0367 and week 4; P = 0.0167) (Fig. 1a).
The HCV RNA log10 decline from baseline to week 1 and week 4 in sustained virological responders, responder/relapsers and nonresponders among genotype 1-infected patients is shown in Fig. 1b. Nonresponders had no or only a limited decline at week 1 and week 4, whereas sustained virological responders had a significant decline both at week 1 and week 4 compared with nonresponders (P = 0.0032 and P = 0.0018, respectively). Also, responder/relapsers had a more pronounced decline at week 1 and week 4 compared with the only nonresponder (P = 0.0367 and P = 0.0167, respectively).
The mean log10 HCV RNA decline in genotype non-1-infected patients was also more pronounced among sustained virological responders than in the only nonresponder at week 1 and week 4; a statistical comparison however, was not judged to be meaningful (Fig. 1c).
Subgroup analyses of HCV RNA kinetics at day 1, week 1 and week 4--
Baseline characteristics of the 15 patients (genotypes 1 and non-1, 9 and 6, respectively) tested for HCV RNA at day 1 in addition to week 1 and week 4 are depicted in Table 2. No significant difference was noted in HCV RNA levels between sustained virological responders and the combined group of responder/relapsers and nonresponders at baseline or day 1, whereas differences were noted at week 1 (P = 0.0265) and week 4 (P = 0.0293), when sustained virological responders had a more pronounced HCV RNA decline. When genotype 1-infected patients with SVR (n = 5) and response/relapse (n = 1) were analysed together and compared with nonresponders (n = 3) at baseline and day 1, no significant differences in HCV RNA levels were noted, whereas significant differences were noted at week 1 and week 4 (0.12 x 106 IU/mL vs 1.8 x 106 IU/mL, P = 0.0201 and 0.0008 x 106 IU/mL vs 0.81 x 106 IU/mL, P = 0.0455, respectively) (Fig. 2a). All patients with genotype non-1 (n = 6) in this subgroup achieved SVR (Fig. 2b).
When patients infected with genotype 1 and non-1 in this subgroup were compared, no significant differences in median HCV RNA levels were noted at baseline and day 1. At weeks 1 and 4, there was a significantly greater decline in HCV RNA levels among genotype non-1-infected patients compared with genotype 1-infected patients (0.29 x 106 IU/mL vs 0.017 x 106 IU/mL, P = 0.0451; and 0.015 x106 IU/mL vs 0.00059 x 106 IU/mL, P = 0.0201, respectively) (Fig. 2c).
When the HCV RNA log10 decline from baseline to day 1 in genotype 1- and genotype non-1-infected patients was compared, no significant difference was noted. However, the decline from baseline to week 1 before the second dose of PEG-IFN was administered was significantly more pronounced in the latter group of patients (P = 0.0292). When the HCV RNA decline in sustained virological responders was compared with the combined group of response/relapsers and nonresponders in this subgroup, no significant difference was observed in HCV RNA decay from baseline to day 1, whereas a trend was seen from baseline to week 1 (P = 0.0583). This might however, have been missed due to dilution of baseline samples which might artificially have increased the log change although all samples were handled according to the instructions of the manufacturer.
HCV RNA kinetics at week 1 and week 4 among all patients--
The sustained virologic responders had significantly lower mean serum HCV RNA levels at week 1 and 4 (0.046 x 106 IU/mL and 0.0097 x 106 IU/mL, respectively) compared with responder/relapsers and nonresponders combined (0.68 x 106 IU/mL and 0.38 x 106 IU/mL; baseline; P = NS, week 1; P < 0.0001 and week 4; P = 0.0004) (Table 3). Sustained virological responders also had a more pronounced HCV RNA decline compared with the combined group of responder/relapsers and nonresponders at weeks 1 and 4 (P = 0.0003 and P = 0.0159, respectively) (Table 3). The decline in HCV RNA levels from weeks 1 to 4 was greater in sustained virological responders compared with responder/relapsers (P = 0.0528), and nonresponders (P = 0.0455).
Log10 HCV RNA decline among all patients according to final outcome--
Week 1:
Twenty-six of the 34 sustained virological responders (76.5%) had an HCV RNA decline of >1 log10 (mean 1.99 log10) at week 1. Eleven of the 34 sustained virological responders (32.3%) had a >2-log10 drop in HCV RNA levels at week 1 compared with only one of 21 (4.8%) patients in the combined group of responder/relapsers and nonresponders, P = 0.0161 (Table 3). In one patient with an SVR (a 42-year-old man infected with genotype 3a), a small increase in HCV RNA (0.25 log10) was noted at week 1. By week 4, a 1.87-log10 decline had occurred, and at week 12, he was HCV RNA negative.
Week 4:
Among sustained virological responders, the HCV RNA decline continued, and at week 4, only four of these individuals had HCV RNA levels above 600 IU/mL (700, 800, 30 000 and 282 000 IU/mL, respectively) (Fig. 1a,b). Thirty-six patients had a >2-log10 drop in HCV RNA levels at week 4. Of these, 29 had genotype non-1 infection (23 responders and six responder/relapsers) and seven had genotype 1 infection (six sustained responders and one responder/relapser).
Week 12:
Only one of 34 (2.9%) sustained virological responders was positive for HCV RNA at week 12 vs 11 of 19 (58%) responder/relapsers, P < 0.0001. The sustained virological responder who was HCV RNA-positive at week 12 was a 55-year-old man infected with HCV genotype 1b who had a minimal HCV RNA decline at weeks 1 and 4 (0.52 log10 and 0.66 log10, respectively).
Predictability of final outcome according to viral decline at weeks 1, 4 and 12:
The positive predictive value (PPV) for a SVR was 92, 77 and 81% at weeks 1, 4 and 12, respectively, when either a >2-log10 drop in HCV RNA at week 1 or 4, or a negative HCV RNA test at week 12 was used as the criterion for response (data not shown). The NPV for a nonsustained response (i.e. response/relapse or nonresponse) was 51%, 61% and 92% at weeks 1, 4 and 12, respectively, when the aforementioned criterion was not met (data not shown). For patients with genotype non-1 and a 2-log10 decline in HCV RNA levels the PPV for a SVR was 89% week 1, 79% week 4, and 79% week 12, whereas the corresponding NPV were 43, 40 and 100% respectively (Table 4).
METHODS
Patients
Patients with chronic hepatitis C who participated in two large international, multicentre studies [either a phase III (5) or an open phase IV study] during 2000-2002 were selected from Swedish centres provided sera had been drawn at baseline and weeks 1, 4 and 12 of treatment, and on day 1 of the phase III study. Eighteen patients were selected from the phase III and 42 from the phase IV study. All patients were treatment-naïve and had elevated alanine aminotransferase (ALT) levels and detectable HCV RNA in serum. A liver biopsy consistent with chronic HCV infection was required for inclusion. All patients were negative for HBsAg and HIV, and lacked evidence of autoimmune and/or metabolic liver diseases. All patients gave written informed consent, and the study was approved by the local ethics committee at each centre.
Treatment
Treatment was given with peginterferon alpha-2a (PEGASYS®; Roche Pharmaceuticals, Nutley, NJ, USA) 180 mug subcutaneously once weekly in combination with ribavirin (COPEGUS®; Roche Laboratories Inc., Nutley, NJ, USA) daily at a dose of 800 or 1000-1200 mg during 24 or 48 weeks, as reported earlier [5], or at a dose of 800-1200 mg daily, depending on body weight (in the phase IV study) during 24 weeks for infections caused by genotype non-1 and 48 weeks for genotype 1. An SVR was defined as the absence of HCV RNA at the end of treatment and follow-up (24 weeks post-treatment); response/relapse was defined as the absence of HCV RNA at the end of treatment and reappearance during follow-up; nonresponse was defined as the presence of HCV RNA during and after treatment.
Methods
Sera were frozen at -70 °C within 2 h of sampling at baseline (day 0), day 1 (in the phase III study), weeks 1, 4 and 12 before the IFN dose was given. All sera were later analysed for HCV RNA levels by the COBAS AMPLIPREP/COBAS Amplicor HCV MONITOR® Test, v2.0 (Roche Diagnostics, Mannheim, Germany), which has a sensitivity of approximately 600 IU/mL. Samples with HCV RNA levels >550 000 IU/mL were retested after dilution according to the instructions from the manufacturer [17]. At the end of treatment and follow-up, a qualitative HCV RNA test was performed by the COBAS Amplicor® HCV Test v2.0 (Roche Diagnostics), which has a sensitivity of 50 IU/mL [18]. HCV genotyping was performed with a line probe assay (Inno-LiPA® HCV II; Innogenetics NV, Gent, Belgium) [19] or an in-house method [20]. Liver biopsy findings were scored according to inflammation (grade) and fibrosis (stage) on a scale of 0-4 [21,22].
Statistics
The statistical evaluation was based on per protocol analysis. HCV RNA levels of <600 IU/mL were set to 599 IU/mL for statistical analyses. The Mann-Whitney U-test was used to test quantitative variables. The Fisher exact two-tailed test was used to test categorical variables. A P-value <0.05 was considered statistically significant. Mean HCV RNA levels were used to describe decrease in HCV RNA levels between different time points, whereas median levels were used in the box plot figures.
|
|
| |
| |
|
|
|