 |
 |
 |
| |
Resistance to Tipranavir & other PIs
|
| |
| |
Excerpted from Resistance Before Therapy, Hidden Mutations, and Blips
46th ICAAC, September 27-30, 2006, San Francisco
Mark Mascolini
How different mutations affect boosted PIs
Analysis of 9860 HIV isolates shipped to Monogram for resistance testing yielded three noteworthy findings about resistance to ritonavir-boosted tipranavir and other PIs [7]:
- The I84V mutation--with or without other key PI mutations--had the biggest impact on all boosted PIs studied, including tipranavir.
- The common V82A mutation--without other critical PI mutations--had no affect on susceptibility to tipranavir and had little effect when joined by a second seminal PI mutation.
- L90M, another common PI mutation, had the least impact on resistance to any PI studied except saquinavir.
Peter Piliero of Boehringer Ingelheim, tipranavir's maker, cautioned that the analysis has limits. Most importantly, the proportion of viral isolates judged susceptible or resistant depends on the Monogram-defined upper clinical cutoff for resistance to tipranavir (8), amprenavir (11.5), saquinavir (12), atazanavir (20), and lopinavir (55). Statisticians controlled the analysis only for major PI mutations assessed in this study--any substitution at protease positions 23, 24, 30, 32, 46, 47, 48, 50, 54, 82 (except V82I), 84, 88, and 90, plus L33I and L33F. (V82I is a mutation selected by tipranavir.) The number of isolates with genotypic and phenotypic data differed for each of the PIs: 9860 isolates had genotypic and phenotypic data for amprenavir, lopinavir, and saquinavir, 9007 for atazanavir, and only 2272 for tipranavir. Finally, Piliero could not reckon susceptibility to a rival PI, darunavir (TMC114).
Nearly 60% of the isolates scrutinized had one or two key PI mutations, and 38.5% had two or three mutations. Two or more of the mutations studied rendered 50% of isolates resistant to amprenavir, 58% resistant to atazanavir, and 70% resistant to saquinavir. Three or more mutations made 70% of isolates resistant to lopinavir, 79% resistant to atazanavir, 82% resistant to saquinavir, and 85% resistant to amprenavir. In contrast only 17% of isolates with two key mutations and 38% with 3 key mutations proved fully resistant to tipranavir.
Table 2 shows the impact of various single and combined mutations on resistance to the PIs studied.
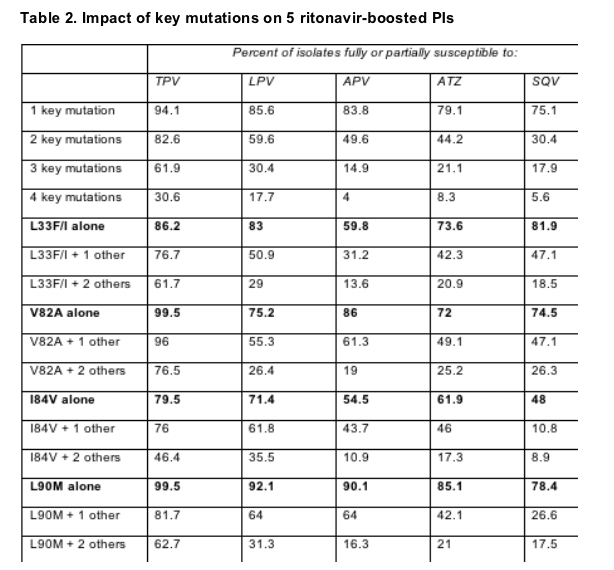
V82A, a mutation that arises with failing atazanavir, indinavir, lopinavir, ritonavir, or saquinavir, did not make HIV resistant to tipranavir, and rarely did so even when accompanied by one or two other key mutations. That makes sense, Piliero observed, because failure of tipranavir selects V82I or V82T, not V82A.
I84V alone had the greatest impact on every one of the PIs studied and, except for saquinavir, L90M had the least impact on these PIs. But L90M plus one other critical mutation made one third to three quarters of isolates resistant to lopinavir, amprenavir, atazanavir, and saquinavir, whereas L90M plus one made fewer than 20% of isolates resistant to tipranavir.
Just after ICAAC, Boehringer and a panel of experts published a list of 21 mutations at 16 protease positions that confer resistance to tipranavir (10V, 13V, 20M/R/V, 33F, 35G, 36I, 43T, 46L, 47V, 54A/M/V, 58E, 69K, 74P, 82L/T, 83D, and 84V).[8] The scoring system that comes with this list calls virus with four or fewer of these mutations susceptible to tipranavir, virus with five to seven partially resistant, and virus with eight or more resistant.
References
7. Piliero PJ, Narkin N, Mayers D. Impact of protease mutations L33F/I, V82A, I84V, and L90M on ritonavir-boosted protease inhibitor susceptibility. 46th ICAAC. September 27-30, 2006. San Francisco. Abstract H-998.
8. Baxter JD, Schapiro JM, Boucher CA, et al. Genotypic changes in human immunodeficiency virus-1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. Virology 2006;80:10794-10801.
|
| |
|
 |
 |
|
|