 |
 |
 |
| |
Sensitive Genotyoic Test Detects Low-Level K65R Which Impairs Viral Response
|
| |
| |
Reported by Jules Levin
XV Intl HIV Drug Resistance Workshop
June 13-17, 2006
Sitges, Spain
This is one of three studies presented at the Workshop raising a concern that sensitive genotypic testing can detect resistance that standard genotpye testing does not detect and that it may affect virologic response.
"Allele-Specific PCR (sensitive genotype test) Shows Low-Level
K65R in Treatment-Experienced Patients with L74V in the Absence of TAMs"
E Svarovskaia, N Margot, A Bae, J Waters,
K Borroto-Esoda, and MD Miller
Gilead Sciences, Inc., Foster City, California, USA
Mike Miller reported these study results and information in an oral presentation at the Workshop.
Author conclusions
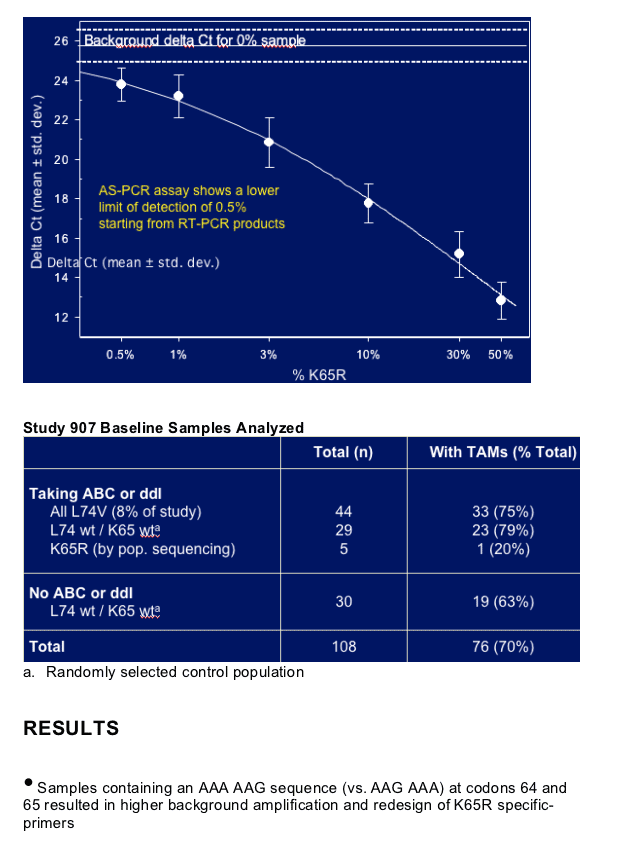
No K65R was detected among >40 treatment-naive patients by an AS-PCR technique (sensitive genotype test) with a lower cut-off of 0.5%
- Strong "A" rich region limits sensitivity of assay at K65
Natural polymorphism at codon 65 (AAG) can interfere with AS-PCR for K65R
- AAG is predominant sequence for subtype C
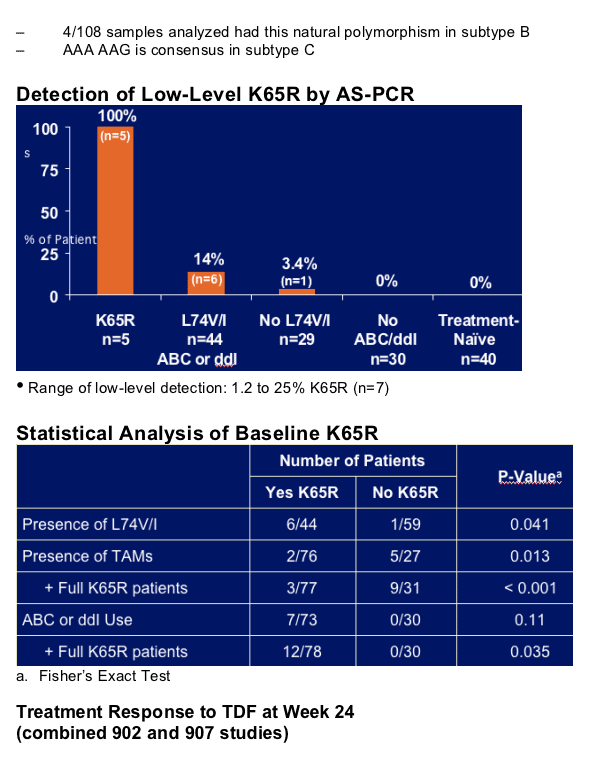
Low-level K65R (1.2% - 25%) detected in 7 of 73 patients (9.6%) pre-treated with ddI or ABC
- Positively associated with L74V and negatively associated with TAMs
- No detection among NRTI-experienced patients not taking ddI or ABC
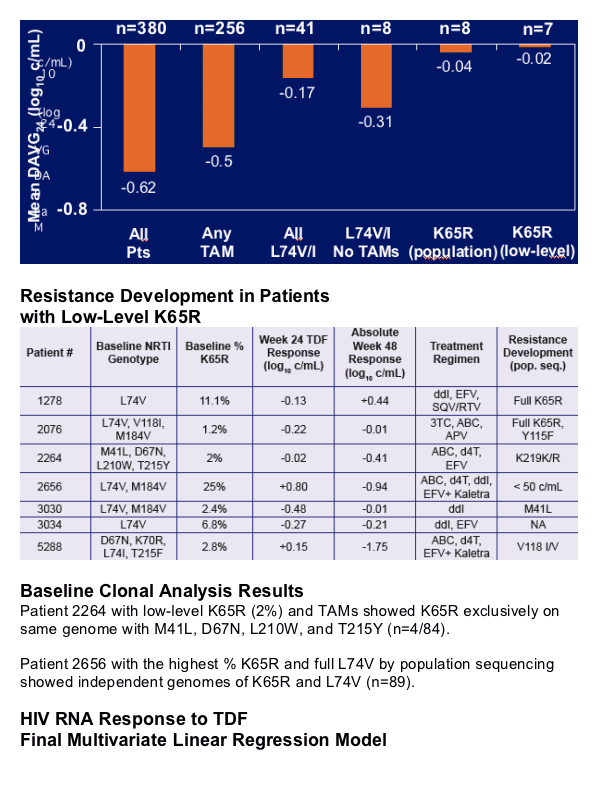
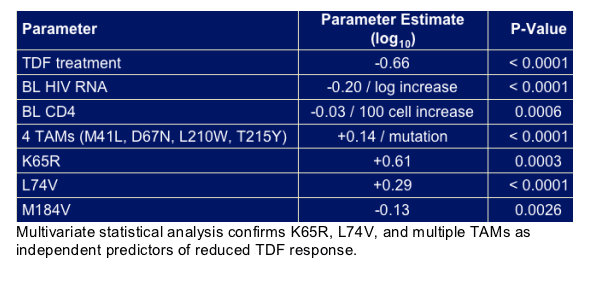
- Reduced treatment responses to TDF with 2 patients developing full K65R
Along with baseline K65R and multiple TAMs, L74V is an independent predictor of reduced response to TDF therapy in heavily treatment-experienced patients
Study Objective
Assess the frequency of low-level K65R in the plasma HIV-1 of treatment-experienced and treatment-naive patients and correlate with response to TDF treatment
Introduction
Prior abacavir or ddI therapy can result in development of either L74V/I or K65R mutations in RT
L74V shows full in vitro susceptibility to tenofovir; however, patients with L74V have shown significantly reduced response to tenofovir DF therapy1
Prior abacavir therapy also predicts poor treatment response to tenofovir DF2
Previous analyses by single-genome sequencing identified two patients in study 907 with L74V/I by population sequencing and low-level K65R at baseline who had poor treatment response and full development of K65R.
Methods
Study 907 was a randomized, placebo-controlled study of tenofovir DF
-- > 8 weeks prior stable background ART to which tenofovir DF or placebo was added
-- HIV RNA > 400 and < 10,000 copies/mL
-- Mean 5.4 years prior ART
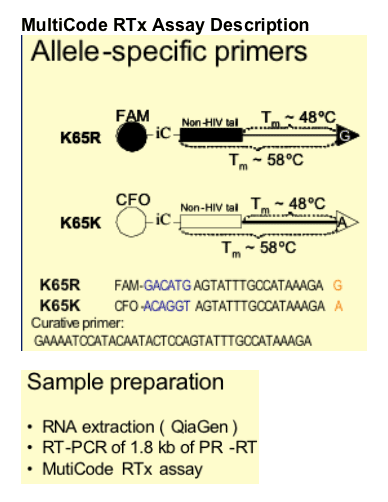
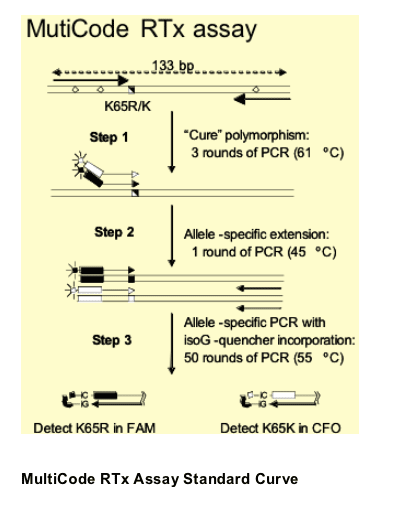
An allele-specific PCR (AS-PCR) assay was developed to detect the K65R mutant using the MultiCode RTx real-time PCR platform (EraGen Biosciences, Madison, WI)
-- For viral loads < 100,000 copies/mL, plasma HIV-1 RNA was pre-amplified by standard procedures
-- Viral standard curve stocks were obtained by electroporation of NL4-3 plasmids into MT2 cells
|
| |
|
 |
 |
|
|