 |
 |
 |
| |
Phenotypic Susceptibility to TMC-114 and Tipranavir Before and After Lopinavir/ritonavir-based Treatment in Subjects Demonstrating Evolution of Lopinavir Resistance
|
| |
| |
Reported by Jules Levin
M King1, TP Young1, B Bernstein1, L Klein1, DK Tokimoto1, MJ Fath1, N Parkin2, GJ Hanna1, DJ Kempf1
1Abbott, Abbott Park, IL; 2Monogram Biosciences, South San Francisco, CA
XV HIV Drug Resistance Workshop
June 13-17, 2006, Sitges, Spain
Abstract #29 poster.
BACKGROUND
- A previous analysis of 275 protease inhibitor-experienced subjects in two Phase 2 and one Phase 3 studies of lopinavir/ritonavir (LPV/r) identified 19 subjects with incremental evolution of LPV resistance during treatment with a LPV/r-based regimen (Mo 2005).
- Subjects with an intermediate level of LPV resistance at study baseline had the highest risk of demonstrating evolution of additional LPV resistance.
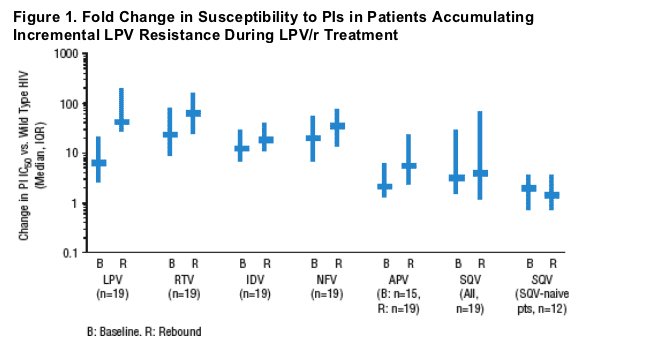
- Viral isolates from subjects with evolution of LPV resistance generally retained or developed a high degree of cross-resistance to ritonavir, indinavir, and nelfinavir, but a lower degree of resistance to saquinavir and amprenavir was observed (Figure 1).
- Changes in resistance to the new protease inhibitors TMC-114 (darunavir) and tipranavir (TPV) during evolution of LPV resistance have not previously been
assessed.
For subjects with inadequate viral response (virologic rebound or failure to achieve undetectable plasma HIV-1 RNA) and no documented interruption of LPV/r, samples from baseline and the time of rebound were retrospectively submitted for genotypic and phenotypic drug resistance testing.
- Incremental evolution of LPV resistance was defined as >2.5-fold reduced susceptibility to LPV in the rebound sample compared to a WT virus (fold change, FC) as well as satisfying one or both of the following criteria:
- Emergence of a new primary mutation in the PR gene associated with PI resistance (D30N, V32I, G48V, I50L/V, V82A/F/T/S, I84V, or L90M)
- Emergence of a new secondary mutation (L10F/I/R/V, K20M/R, L24I, L33F, M36I, M46I/L, I47A/V, I54A/V/L/S, A71V/T, G73S/A, V77I, or N88D) accompanied by an increase greater than or equal to twofold in the LPV IC50 between baseline (pre-LPV/r treatment) and rebound.
- For the current analysis, subjects with evolution of LPV resistance and available archive samples had samples submitted for testing of phenotypic susceptibility to
TMC-114 and tipranavir.
- TMC-114 was provided by Abbott to Monogram Biosciences in a blinded fashion.
- Viral susceptibility and replication capacity were determined by Monogram Biosciences using PhenoSense (Petropoulos 2000, Campbell 2003).
RESULTS
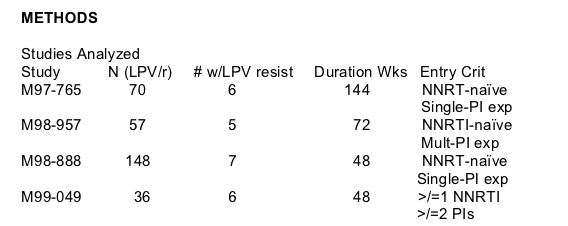
18 subjects with available samples met inclusion criteria and had phenotypic resistance testing conducted (Figure 2) on samples prior to LPV/r-treatment
("baseline") and at virologic rebound during LPV/r treatment ("rebound"). No subject demonstrated 2.5 FC in LPV susceptibility between baseline and rebound
without accompanying primary or secondary mutations.
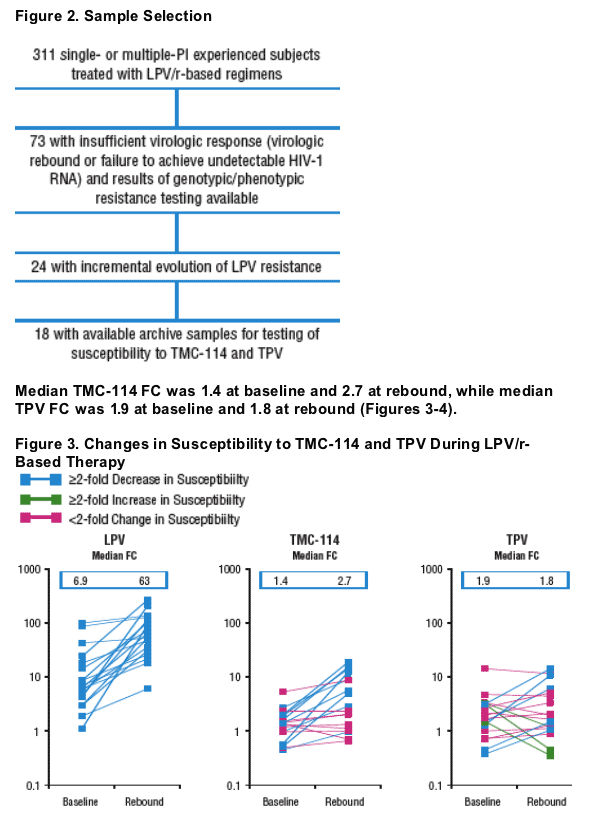
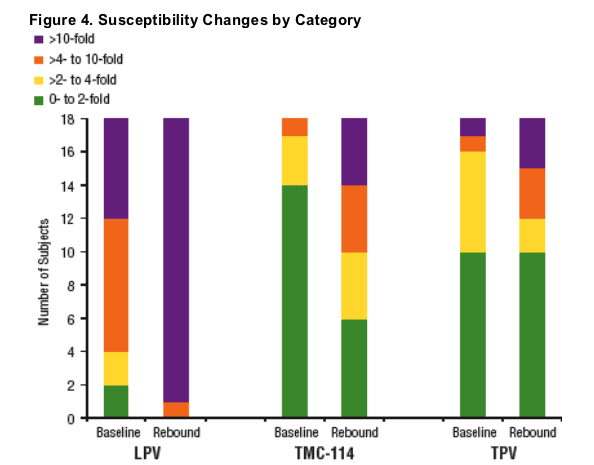
Isolates from 9 subjects demonstrated 2-fold decrease in TMC-114 susceptibility from baseline to rebound.
Isolates from 5 subjects demonstrated 2-fold decrease, whereas isolates from 3 subjects demonstrated 2-fold increase in TPV susceptibility from baseline
to rebound.
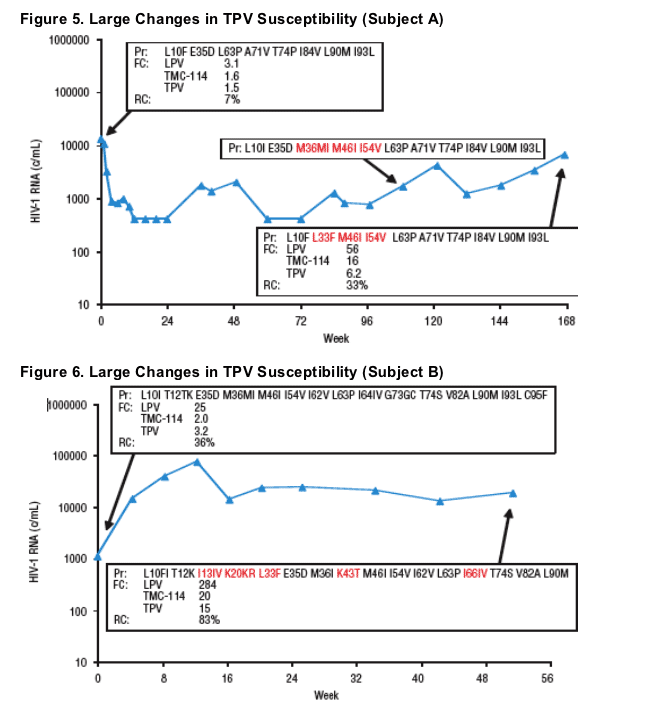
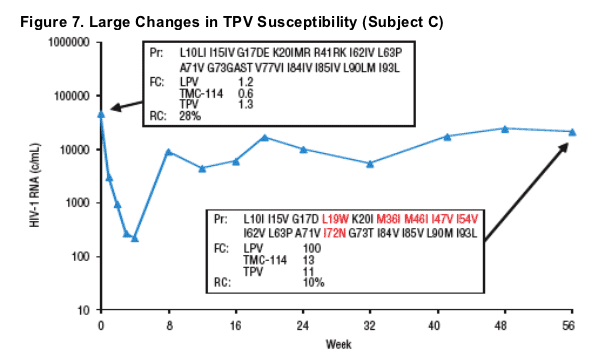
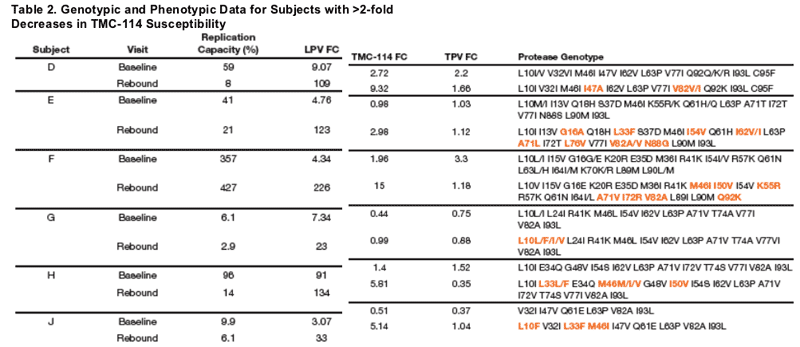
Plasma HIV-1 RNA, genotype, and phenotype data for 3 subjects with >2-fold decrease in TPV susceptibility and rebound TPV FC >2-fold are displayed in Figures 5-7. Baseline and rebound data for the other 6 subjects with >2-fold increase in TMC-114 susceptibility are shown in Table 2.
- Two of these 3 subjects demonstrated emergence of the L33F mutation at rebound.
- All 3 subjects had mutations L10F/I, L90M, and either V82A or I84V at baseline, and all either had M46I and I54V mutations at baseline (n=2) or developed them at rebound (n=1).
The emergence of an I50V mutation was observed in isolates from two subjects, with a corresponding 4- to 8-fold decreased susceptibility to TMC-114 (compared
to baseline) but 3- to 4-fold increased susceptibility to TPV, and reductions in replication capacity of >85%. Increases in TPV susceptibility have previously been associated with the I50V mutation (Kohlbrenner 2004). Rebound isolates from the remaining subject with an increase in TPV susceptibility demonstrated emergence of mutations L24I, M46I, and F53F/L with decrease in RC of >90%.
Notably, 6 of the 9 subjects with >2-fold decreases in TMC-114 susceptibility (Figures 5-7 and Table 2) demonstrated the emergence of substitutions different from those associated with TMC-114 resistance to date (V11I, V32I, L33F, I47V, I50V, I54L/M, G73S, L76V, I84V, I89V [de Meyer 2006]), highlighting the potential value of phenotypic resistance testing, especially for new compounds.
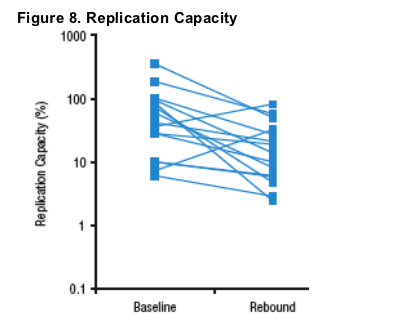
- Replication capacity decreased by a median of 64% (interquartile range: 40-86%) between baseline and rebound (n=17, Figure 8).
- Rebound isolates from 2 subjects with increases in RC both demonstrated emergence of the L33F mutation (Figures 5-6), although rebound isolates from 2 other subjects with emergence of L33F demonstrated 40-50% decreases in RC (Table 2).
The authors concluded:
- Evolution of lopinavir resistance during lopinavir/ritonavir-based treatment in protease inhibitor-experienced subjects was infrequently identified; additional
resistance was observed in 8% of subjects treated for a median of 48 weeks.
- Evolution of incremental resistance to lopinavir/ritonavir generally resulted in lower replication capacity with relatively little change in susceptibility to tipranavir
and modest changes in susceptibility to TMC-114 in some subjects.
- Tipranavir or TMC-114, guided by combination genotype/phenotype resistance testing, may be useful for salvage therapy following evolution of resistance on
a lopinavir/ritonavir-based regimen.
|
| |
|
 |
 |
|
|