 |
 |
 |
| |
Darunavir-Amprenavir Cross-Resistance in Clinical Samples Submitted for Phenotype-Genotype Combination Resistance Testing
|
| |
| |
Reported by Jules Levin
Neil Parkin, Eric Stawiski, Colombe Chappey, and Eoin Coakley
Monogram Biosciences, South San Francisco, CA USA.
14th CROI, Los Angeles, CA, February 25-28, 2007
AUTHOR CONCLUSIONS:
DRV and APV in vitro susceptibility patterns are very similar.
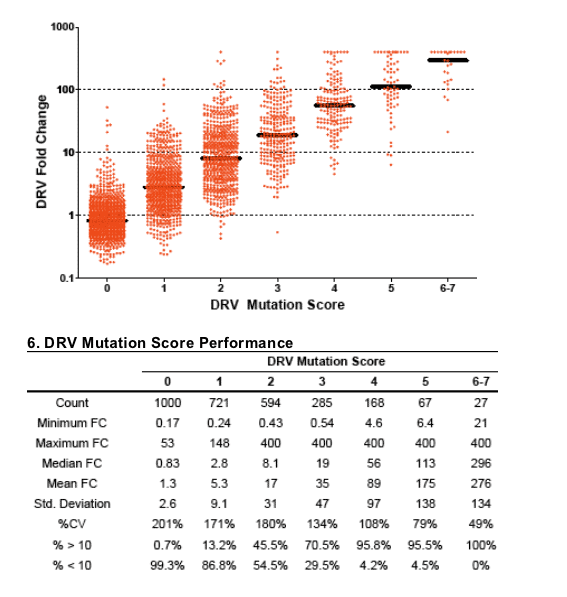
Predicted incidence of clinically meaningful cross-resistance is low, due to differences in CCO which are higher for DRV (see Panel 4). Clinical trial data are
consistent with this prediction (see Picchio et al poster #609).
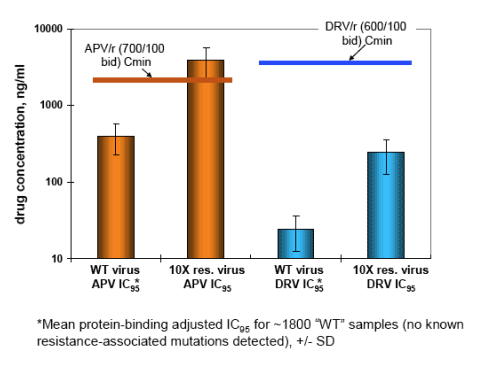
The expected increased efficacy of DRV compared to APV in PI-experienced patients is most likely a result of higher potency (16-fold lower IC50 in the PhenoSense assay) and about 2-fold higher free drug levels in plasma (see Panel 9), rather than a unique cross-resistance profile.
BACKGROUND
Darunavir (DRV, TMC-114) is the most recently approved protease inhibitor (PI).
Mutations correlated with reduced clinical response to DRV* are similar to those known to confer reduced susceptibility to amprenavir (APV).
We examined PI cross-resistance patterns and predictive accuracy of a simple DRV mutation score genotype interpretation among samples tested in the Monogram Clinical Reference Laboratory.
METHODS
Fold-change in IC50 (FC) data from 2862 samples tested since May 2006 (PhenoSense HIV, Monogram Biosciences) with at least one major PI mutation and no mixtures at a position included in the DRV mutation score were analyzed.
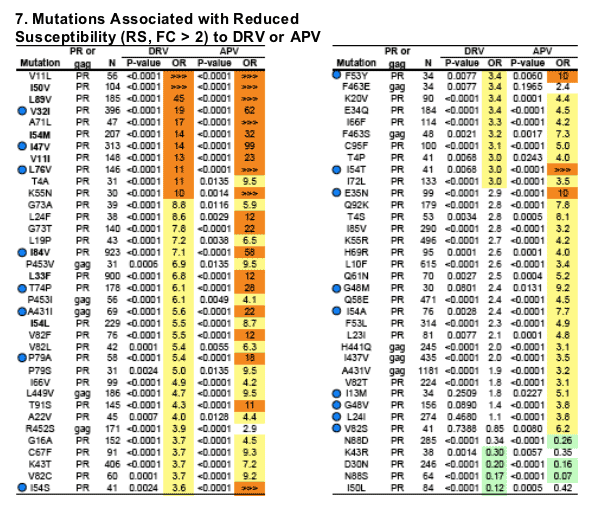
The DRV mutation score was calculated as the number of mutations present among V11I, V32I, L33F, I47V, I50V, I54L or M, G73S, L76V, I84V, and L89V*; the presence of 3 or more of these mutations was considered to indicate reduced susceptibility.
Samples with DRV IC50 greater than the highest drug concentration tested were assigned an arbitrary maximum FC value of 400.
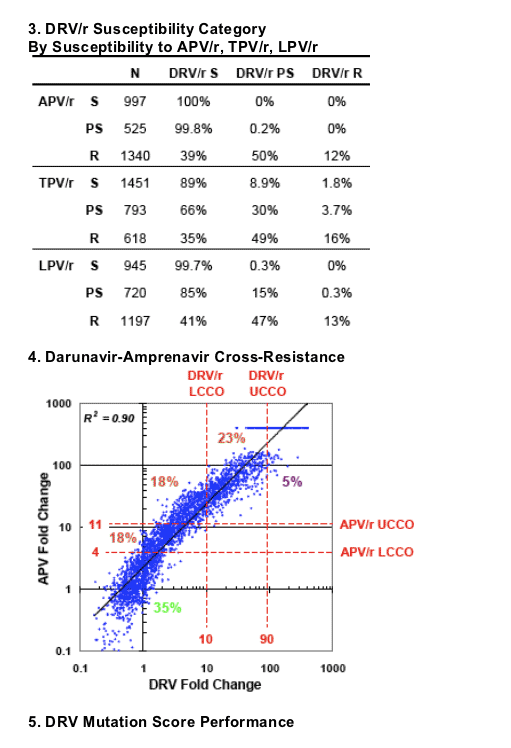
Samples were classified as drug sensitive (S), partially sensitive (PS), or resistant (R) based on lower and upper clinical cutoffs (CCO) (see Coakley et al, poster #610).
Mutations in gag (aa 418-500) or protease associated with reduced susceptibility (RS, FC>2) were determined using Fisher's Exact test with correction for multiple comparisons.
RESULTS
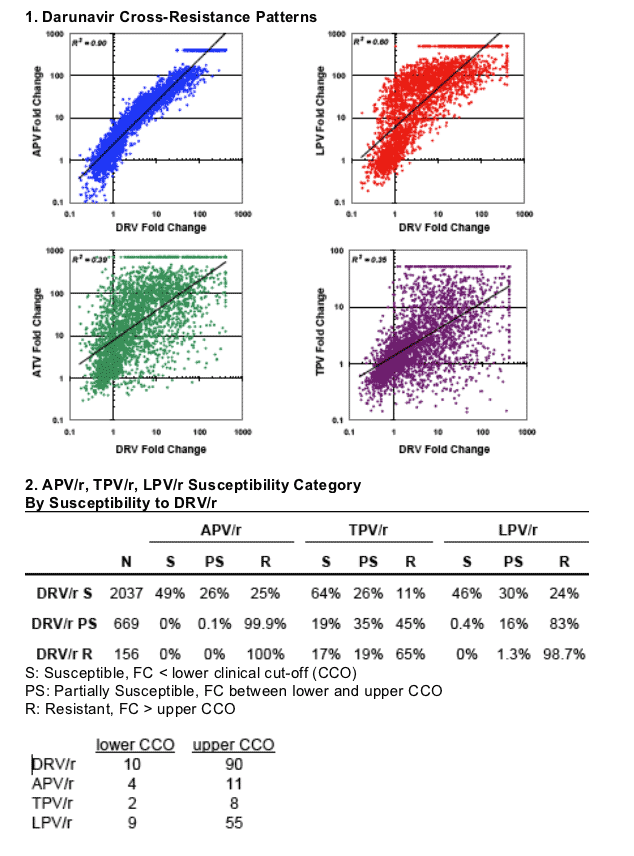
The coefficient of correlation R2 for log-transformed PI FC data was highest between DRV and APV (0.90), intermediate for lopinavir (0.60), and relatively low for other PIs (range 0.25-0.51) (Panel 1).
All 156 DRV/r resistant samples were also resistant to APV/r; 65% and 98.7% were resistant to TPV/r and LPV-R, respectively (Panel 2).
524 of 525 APV/r partially susceptible samples retained DRV/r susceptibility, and only 12% of 1340 APV/r resistant samples were resistant to DRV/r; 50% were partially susceptible to DRV/r (Panel 3).
The percentage of samples with 1 (n=721) or 2 (n=594) DRV mutations which were not DRV/r susceptible was 13.2% and 45%, respectively. Conversely, 29% of samples with 3 mutations (n=285) were DRV/r susceptible (Panels 5 and 6).
Of the 72 most significant mutations for either drug (arbitrarily defined as P<0.05 and odds ratio >3 or < 0.3), 68 are significant for APV, 50 for DRV, and 46 for both (Panel 7).
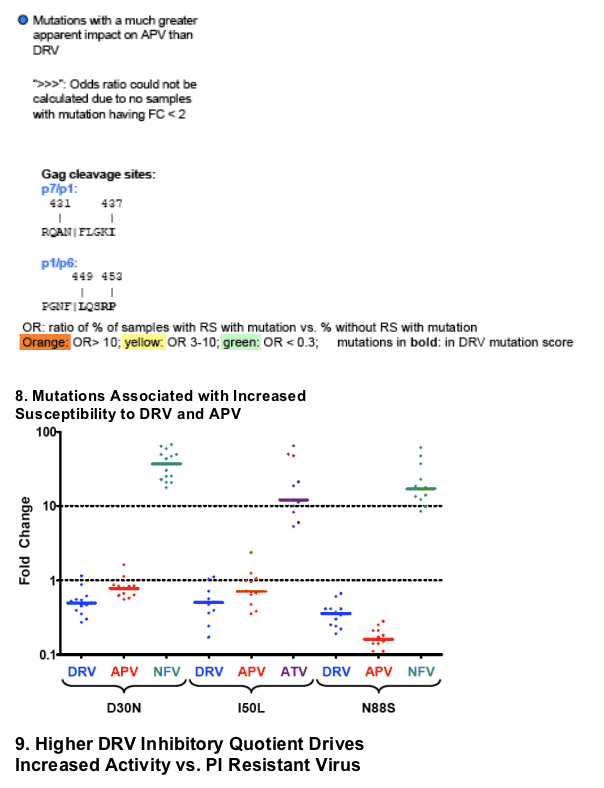
When present as the only major PI mutation, D30N (n=14), I50L (n=11), and N88S (n=12) were associated with increased susceptibility to DRV (median FC 0.49, 0.50, 0.36, respectively; corresponding median APV FC 0.77, 0.71, 0.16) (Panel 8).
PR genotypes of the 7 outliers with DRV mutation score = 0 and DRV FC > 10:
--L10I, I13V, K20T, L33I, E35N/S, M36I, N37D, M46I, I62V, L63P, A71V, V77I, V82F, N88G, L90M
--L10V, T12P, I13V, K14R, K20R, L33I, E35D, M36I, N37E, K43K/T, M46L, G48V, I54T, R57K, I62V, L63P, I64V, I72M, T74S, V77I, V82A
--L10I/V, T12P, K14R, K20R, E35D, M36V, N37T, K43T, G48Q, I54S, K55T, R57R/K, D60E, I62V, L63P/T, A71V, V77I, V82F, L90M, I93L
--L10I, K20R, E35D, M36I, R41K, M46I, I54V, Q58E, I62V, L63A/T, A71V, I72V, V77V/I, V82I, I84A, G86G/R, I93L
--L10I/V, I13I/V, I15V, K20T, E35D, M36I, R41S, M46I, I62V, L63P, A71L, V77I, V82F, N88G, L90M, I93L
--L10V, T12P, K14R, K20R, E35D, M36V, N37T, K43T, G48Q, I54S, K55T, D60E, I62V, L63P, A71V, V77I, V82F, L90M, I93L
--L10I, K20G/R, L24I, E35D, M36V, N37T, R41K, K43T, M46L, R57R/K, I62V, L63P, I72I/V, V77I, V82F, I84C
|
| |
|
 |
 |
|
|