| |
FDA analysis of tipranavir clinical resistance in HIV-1-infected treatment-experienced patients
|
| |
| |
AIDS: Volume 21(2) 11 January 2007 p 179-18
Naeger, Lisa K; Struble, Kimberly A
From the Division of Antiviral Products, Center for New Drug Evaluation, Food and Drug Administration, Silver Spring, Maryland, USA.
".....The use of other active agents with APTIVUS/ritonavir is associated with a greater likelihood of treatment response....Genotypic or phenotypic testing and/or treatment history should guide the use of APTIVUS/ritonavir. The number of baseline primary protease inhibitor mutations affects the virologic response to APTIVUS/ritonavir...."
Abstract
Objective: To assess the resistance profile of tipranavir.
Methods: Resistance analyses were performed on Boehringer Ingelheim-sponsored studies examining the safety and efficacy of tipranavir in highly treatment-experienced individuals at 24 weeks. Virologic response rates based on the presence of baseline primary protease inhibitor mutations and based on baseline tipranavir susceptibility were evaluated, and the development of protease mutations during treatment with tipranavir was analyzed.
Results: Virologic response rates in tipranavir-treated individuals were reduced when isolates with substitutions at amino acid positions I13, V32, M36, I47, Q58, D60 V82 or I84 were present at baseline. In addition, virologic response rates to tipranavir decreased when the number of baseline protease inhibitor (PI) mutations was five or more. Individuals who received tipranavir without concomitant enfurvitide and had five or more baseline PI mutations group began to lose antiviral response between weeks 4 and 8. However, individuals taking enfuvirtide with tipranavir were able to achieve greater than 1.5 log10 reductions in viral load from baseline out to 24 weeks even if they had five or more baseline PI mutations. Virologic response rates to tipranavir decreased when the baseline phenotype for tipranavir had a greater than three-fold shift in the 50% effective concentration (EC50) from reference. The most common protease mutations that developed in tipranavir-treated individuals who experienced virologic failure were L10I/V/S, I13V, L33V/I/F, M36V/I/L V82T, V82L, and I84V. The resistance profile in treatment-naive individuals was not characterized.
Conclusions: Baseline genotypic and phenotypic data provide valuable information on the likelihood of a virologic response to tipranavir.
Introduction
Currently more than 20 antiretroviral drug products are approved in the United States by the Food and Drug Administration (FDA) for the treatment of HIV infection, some in multiple formulations and fixed drug combinations. Unfortunately, HIV develops resistance to antiretroviral drugs over time, frequently from the accumulation of multiple mutations. Therefore, resistance testing is important in clinical trials to identify the baseline genotypic and phenotypic determinants of virologic success or failure and to determine the effect of antiviral drugs on virus evolution. Characterization of resistance/cross-resistance is a critical part of antiretroviral drug development and clinical resistance data should be available at time of approval [1]. In the review of new drug applications for the treatment of HIV, the Division of Antiviral Products at the FDA independently analyzes resistance data submitted from applicants in order to characterize a new antiretroviral drug's resistance profile. The analyses include examination of the development of mutations on treatment, virologic response based on baseline genotype/phenotype, and cross-resistance to other approved antiretroviral drugs. This assessment provides information on the potential for resistance development to a drug and the likelihood of a virologic response to an antiretroviral drug. To efficiently characterize the resistance profile of a new drug, the FDA believes resistance testing should be included in all phases of drug development with an emphasis on early stages of development.
Tipranavir (TPV), developed by Boehringer Ingelheim (Ridgefield, Connecticut, USA), is a novel protease inhibitor (PI) with antiviral activity against multi-PI-resistant clinical HIV-1 isolates with an average 50% effective concentration (EC50) value against these isolates of 240 nmol/l (range 50 to 380 nmol/l) [2-4]. TPV-resistant viruses were selected in vitro when wild-type HIV-1NL4-3 was serially passaged in the presence of increasing concentrations of TPV in cell culture. HIV-1 variants with 70-fold decreased susceptibility to TPV were selected after 9 months in passage, which had 10 mutations arising in this order: L33F, I84V, K45I, I13V, V32I, V82L, M36I, A71V, L10F, and I54V [5]. Mutations in the CA/P2 protease cleavage site and the transframe region were also detected. TPV-resistant viruses showed decreased susceptibility to all currently available protease inhibitors except saquinavir, which had a 2.5-fold reduced susceptibility [5].
The efficacy of ritonavir-boosted tipranavir (TPV/r) was examined in highly treatment-experienced HIV-infected patients in two Phase III trials [6]. Analyses were conducted to evaluate HIV-1 RNA response according to the presence and absence of baseline protease mutations [7]. These analyses helped assess the association between a specific mutation or mutational pattern and the potential for a virologic response to the drug. Response rates were also examined by baseline susceptibility to TPV (phenotype). In addition, protease mutations that developed during treatment with TPV/r were examined. These analyses provide valuable information for clinicians and HIV patients so they can use TPV/r to its maximal clinical benefit.
Methods
Study population and baseline characteristics
Two identically designed trials, RESIST 1 (1182.12) and RESIST 2 (1182.48), sponsored by Boehringer Ingelheim were multi-center, multi-national, randomized, controlled, open-label studies in HIV-infected individuals with triple antiretroviral class and dual PI-drug regimen experience. RESIST 1 was conducted in the United States, Canada and Australia, while RESIST 2 was conducted in Europe and Latin America. Participants were randomized to receive either TPV/r 500 mg/200 mg administered twice daily (n = 582) or an investigator-selected comparative protease inhibitor boosted with ritonavir (CPI/r) (n = 577) in combination with an optimized background regimen (OBR). In RESIST 1 and 2, the safety and efficacy of TPV/r and CPI/r through 24 weeks of treatment were compared [6].
Genotypic resistance testing was performed at screening to enroll individuals with at least one primary PI mutation at codons 30N, 46I/L, 48V, 50V, 82A/F/L/T, 84V, or 90M and no more than two protease mutations at positions 33, 82, 84, or 90. Resistance data from Phase II Studies such as 1182.052, a study of multiple PI-experienced triple antiretroviral class-experienced individuals on 2-week functional TPV/r monotherapy followed by 30 weeks of TPV/r plus optimized antiretroviral therapy, was used to formulate the exclusion criterion of no more than two protease mutations at positions 33, 82, 84, or 90 for the RESIST trials [8].
The CPI/r regimen was selected based on genotypic resistance results from the TRUGENE (Bayer Healthcare, Tarrytown, New York, USA) and Virtual Phenotype (Tibotec, Mechelen, Belgium) assays. Investigators selected optimized background regimens based on genotypic data and participants were randomized equally to either the TPV/r or CPI/r arm and stratified by pre-selected PI and use of enfuvirtide (ENF). Due to the complex comparator treatment group containing various ritonavir-boosted protease inhibitors with varying degrees of baseline resistance to comparator drugs, the studies were superiority open-label trials.
The patient populations in RESIST 1 and 2 were highly treatment-experienced with a median viral load of 4.82, median CD4+ cell counts of 155, and a median number of four (range one to seven) PIs received prior to study. In the combined RESIST trials at baseline, 97% of the isolates were resistant to at least one PI, 95% of the isolates were resistant to at least one nucleoside reverse transcriptase inhibitor (NRTI), and > 75% of the isolates were resistant to at least one non-nucleoside reverse transcriptase inhibitor (NNRTI). The treatment arms from both studies were balanced with respect to baseline genotypic and phenotypic resistance with 30% of the isolates resistant to TPV at baseline and 80-90% of the isolates resistant to PIs - amprenavir, atazanavir, indinavir, lopinavir, nelfinavir, ritonavir or saquinavir. The median number of baseline PI, NRTI, NNRTI mutations was equivalent between the TPV/r and CPI/r arms in both studies. At baseline, the median number of FDA-defined primary PI mutations (D30, V32, M36, M46, I47, G48, I50, I54, F53, V82, I84, N88 and L90) was four and the median number of IAS protease mutations (amino acid positions 10, 20, 24, 30, 32, 33, 36, 46, 47, 48, 50, 53, 54, 63, 71, 73, 77, 82, 84, 88, or 90) was nine. The median number of NNRTI and NRTI mutations at baseline was one and five, respectively.
Genotypic and phenotypic methods
Genotypes from 1482 isolates were submitted for review. Genotypes were determined by two different assays in RESIST 1 and 2. In RESIST 1, the TRUGENE assay version 1.0 was used. If a sample could not be amplified and genotyped, version 1.5 was used. In RESIST 2, the Virtual Phenotype assay was used for samples from Europe and the TRUGENE assay was used for samples from Latin America and Australia. Genotypic resistance testing was used to stratify participants by pre-selected protease inhibitors (amprenavir, indinavir, lopinavir, saquinavir). For the purpose of stratification, protease inhibitor sensitivity was interpreted from genotypic reports as not resistant, possibly resistant or resistant.
The Virco Antivirogram (Tibotec) assay was used to determine phenotype for virologic failure samples and for randomly selected baseline samples from the RESIST trials (n = 454; 361 from the TPV/r arm and 93 from the CPI/r arm).
Methods of analysis
Baseline resistance analyses were conducted on a censored patient population to assess the impact of baseline resistance and outcome without confounding factors such as early discontinuation due to adverse events [7]. Participants who discontinued study treatment while suppressed or who discontinued study treatment before confirmed suppression for adverse event, non-compliance, protocol violation, pregnancy, or withdrew consent were censored. In addition, participants were censored if they did not reach week 24, had no week 8-24 HIV-1 RNA data, had HIV-1 RNA data only up to week 8 and achieved at least 0.5 log10 decrease, or added a new antiretroviral or changed PI. This approach for analyzing baseline resistance data is consistent with the FDA Draft Guidance for Industry: 'Role of HIV Drug Resistance Testing in Antiretroviral Drug Development' and the FDA analyses for other antiretroviral drugs [1]. Following these criteria, 467 individuals were censored providing a resistance dataset of 1015 participants for analysis of the primary endpoint (≥ 1 log10 decrease in HIV-1 RNA from baseline confirmed). The majority of individuals censored in the analyses were from RESIST 2 because only the week 16 data were available for review. For analysis of time-averaged change from baseline at week 24 (DAVG24), individuals from RESIST 2 who did not reach week 24 but had week 16 data available were included, which provided a larger dataset of 1409 participants.
To assess virologic outcome, several endpoints including the proportion of responders with confirmed ≥ 1 log10 decrease at week 24, DAVG24, and median change from baseline at weeks 2, 4, 8, 16, and 24 were evaluated. In addition, because individuals were stratified based on ENF use, virologic outcomes were examined in three groups: overall (All), those not receiving ENF (No ENF), and those receiving ENF (+ENF) as part of OBR. Many of the analyses were focused on the No ENF group in order to assess baseline resistance predictors of TPV/r virologic outcome without the additive effect of ENF use on the overall response.
Results
Baseline genotype and virologic response analyses
An analysis was conducted to assess virologic response rates by the number of baseline PI mutations using week 24 endpoints of a confirmed greater than or equal to 1 log10 HIV RNA decrease from baseline and a time-averaged change in HIV-1 RNA from baseline (DAVG24). In this analysis, any substitution at protease amino acid positions D30, V32, M36, M46, I47, G48, I50, I54, F53, V82, I84, N88 and L90 was counted if present at baseline. These mutations were selected from several resources and are associated with reduced susceptibility to currently approved PIs [9-11].
In both the TPV/r and CPI/r arms, response rates were similar to or greater than the overall response rates for the respective treatment groups for individuals with one to four PI mutations at baseline (Table 1). Response rates were reduced if five or more PI-associated mutations were present at baseline. Of the individuals who did not use ENF in their OBR, 28% in the TPV/r arm and 11% in the CPI/r arm had a confirmed 1 log10 decrease at week 24 if they had five or more PI mutations in their HIV at baseline (Table 1). The participants with five or more PI mutations in their HIV at baseline without ENF in their OBR achieved a 0.86 log10 median DAVG24 decrease in viral load on TPV/r treatment (n = 215) compared with a 0.23 log10 median DAVG24 decrease in viral load on CPI/r treatment (n = 258) (data not shown). However, participants with five or more baseline PI mutations in their HIV who received TPV/r and ENF (n = 88) had a median time averaged reduction (DAVG) in HIV RNA of 1.88 log10 at week 24 (data not shown). In general, regardless of the number of baseline PI mutations or ENF use, the TPV/r arm had approximately 20% more responders by the primary endpoint (Table 1) and greater declines in viral load by median DAVG24 (data not shown) than the CPI/r arm.
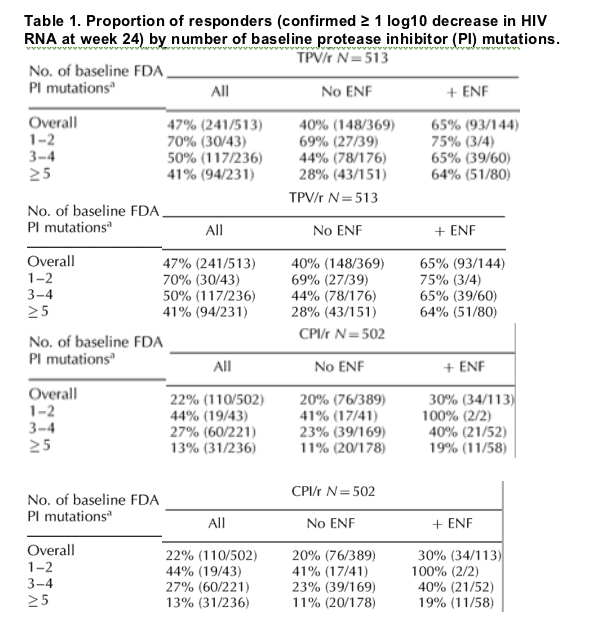
An examination of the median change from baseline of HIV RNA at weeks 2, 4, 8, 16 and 24 by number of baseline PI mutations (1-4 and 5+) showed an approximate 1.5 log10 decline in viral load by weeks 2-4 in the TPV/r arms regardless of the number of baseline PI mutations (1-4 or 5+) (Fig. 1). Individuals who received TPV/r without ENF and who had five or more baseline PI mutations began to lose their antiviral response between weeks 4 and 8 (Fig. 1b). However, sustained viral load decreases of 1.5-2 log10 up to week 24 were observed in individuals receiving TPV/r and ENF (Fig. 1c).
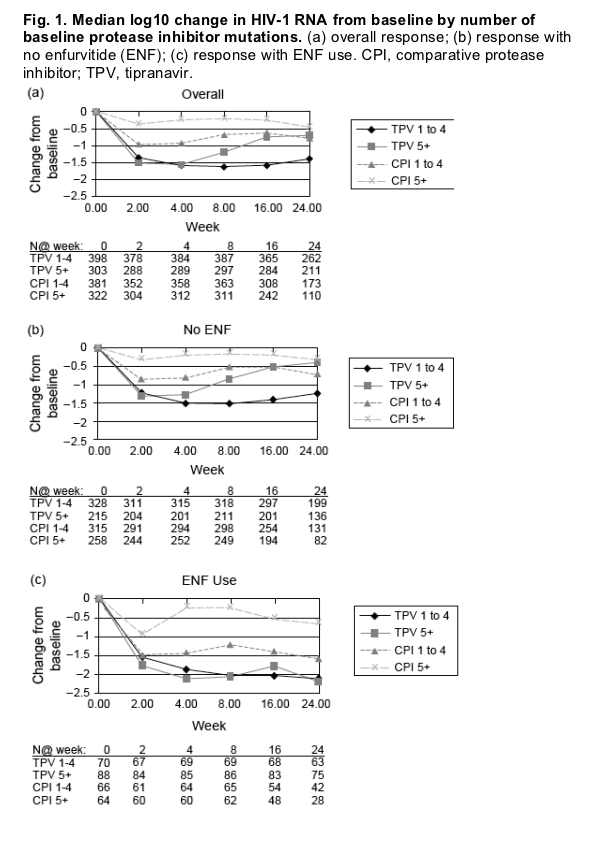
Participants with one to four PI mutations in the CPI/r arm had approximately 1 log10 median decreases in viral load (Fig. 1) unless they were also receiving ENF wherein they obtained approximately 1.5 log10 median decreases in viral load (Fig. 1c). Participants with five or more PI mutations in the CPI/r arm had only an approximately 0.5 log10 viral load decrease even when they received concomitant ENF.
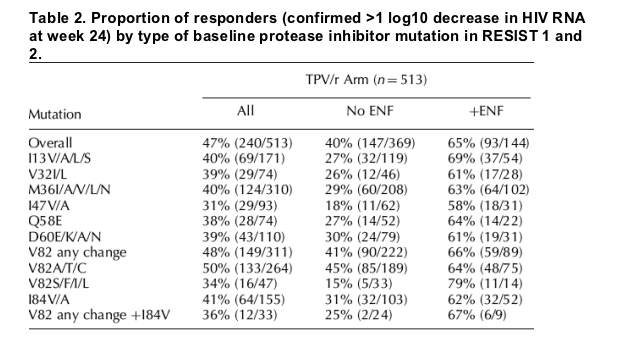
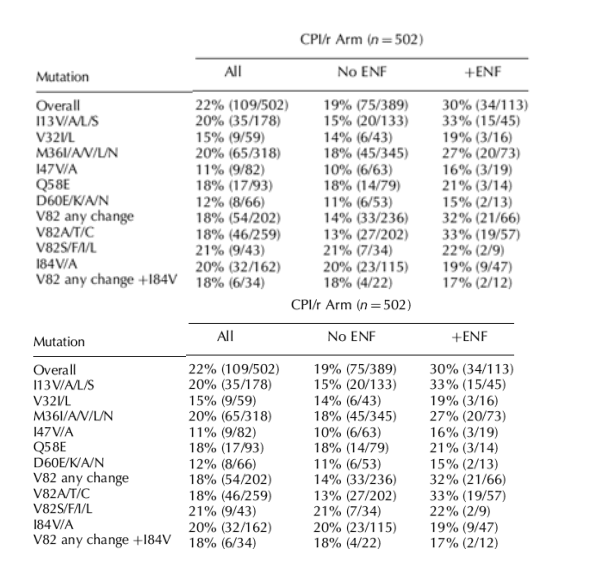
Virologic response rates were also analyzed by the presence of baseline substitutions at each of 25 different protease amino acid positions. Reduced virologic responses were seen in TPV/r-treated individuals when isolates had a baseline amino acid substitution at positions I13, V32, M36, I47, Q58, D60 or I84 (Table 2). Virologic responses were similar or greater than the overall responses when these amino acids were wild type (data not shown). The mutations identified here in the analysis are included in Boehringer Ingelheim's 'Tipranavir score', which was developed by Boehringer Ingelheim to identify protease mutations associated with decreased response to TPV/r [12,13]. The reduction in virologic responses for these baseline substitutions was most prominent in the No ENF subgroup. Responses for viruses with substitutions at position V82 varied depending on the amino acid substitution. Substitutions V82S or F or I or L, but not A or T or C, had reduced virologic responses (15% without ENF use) compared to the overall response (Table 2). Individuals with the I84V mutation and any substitution at V82 including V82A or T had lower response rates than the overall response (25% without ENF use). As mentioned above, the CPI/r arm had approximately 20% fewer responders than the TPV/r arm.
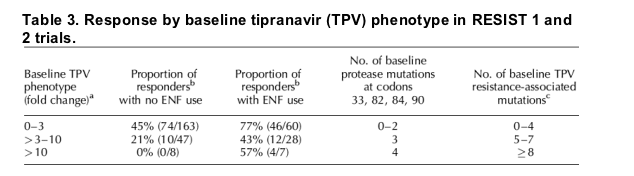
Proportion of responders by baseline TPV phenotype
TPV/r response rates were also assessed by baseline TPV phenotype. The No ENF group was a particular focus in order to more accurately assess the effect of baseline phenotype on virologic outcome to TPV/r. For participants receiving TPV/r without ENF use, the proportion of responders was 45% if the shift in EC50 value from reference of TPV susceptibility was three-fold or less at baseline (Table 3). The proportion of responders decreased to 21% when the TPV baseline phenotype values were between three and ten-fold and 0% when TPV baseline phenotype values were greater than ten-fold. In participants not receiving ENF with TPV/r, the time averaged change in HIV-1 RNA from baseline at week 24 (DAVG24) was -1.31 log10 if the shift in TPV susceptibility was three-fold or less at baseline, -0.41 log10 when the TPV baseline phenotype values were between three and ten-fold, and -0.24 log10 when values were greater than ten-fold (data not shown).
Mutations developing on TPV/r treatment
Baseline and failure isolates were analyzed from treatment-experienced individuals in Phase II Study 1182.052 (n = 32) and the RESIST trials (n = 59) who experienced virologic failure on TPV/r treatment. In this analysis, all patients who failed or discontinued treatment were analyzed with no censoring. The most common mutations that developed in greater than 20% of these TPV/r virologic failure isolates were L33V/I/F, V82T and I84V. Other mutations that developed in 10 to 20% of the TPV/r virologic failure isolates included L10V/I/S, I13V, E35D/G/N, I47V, K55R, V82L and L89V/M/W. The protease mutations that developed in clinical isolates from TPV/r-treated participants also arose in serial in-vitro passage experiments [5].
The V82T mutation developed frequently (34%) in individuals with virologic failure especially when the V82A mutation was present at baseline, whereas isolates with wild-type V82 most often developed V82L. An alanine codon (GCX) at position V82 requires only one change to become threonine (ACX), whereas the wild-type valine codon (GUX) requires two changes to become threonine, but only one to become leucine (CUX). Isolates that developed changes at V82 also frequently developed the I84V mutation: 20% (12/59) of the virologic failures developed both a change at V82 and an I84V mutation.
In RESIST 1 and 2, TPV/r resistance developed in individuals with virologic failure (n = 59) at an average of 38 weeks with a median decrease of 14-fold in TPV susceptibility. The baseline mean TPV susceptibility of the virologic failure isolates was 3.3-fold. These TPV/r resistant isolates were highly cross-resistant to other PIs. Importantly, the resistance profile in treatment-naive individuals has not been characterized.
Discussion
Resistance testing is an important tool in the clinical management of individuals with HIV infection and a critical component in antiretroviral drug development. Resistance analyses in clinical trials aid in identifying the baseline genotypic and phenotypic determinants of virologic success or failure and in determining the effect of an antiviral drug on the evolution of the virus. Several types of resistance analyses were undertaken during the review of tipranavir in order to characterize its resistance profile. The available HIV resistance data provided by the applicant was used during the review in order to examine the virologic response to TPV/r based on baseline genotype/phenotype and the development of HIV protease mutations on TPV/r treatment.
The effect of baseline genotype on the virologic response to TPV was examined in the RESIST trials. The analyses showed both the number and type of baseline PI mutations affected response rates to TPV/r. Virologic response rates in TPV/r-treated participants were reduced when isolates with amino acid substitutions at positions I13, V32, M36, I47, Q58, D60, I84 or substitutions V82S/F/I/L were present at baseline. Furthermore, virologic responses to TPV/r at week 24 decreased when the number of baseline FDA-defined primary PI mutations was five or more. However, individuals receiving ENF with TPV/r were able to achieve > 1.5 log10 reductions in viral load from baseline out to 24 weeks even with five or more baseline PI mutations. The other drugs in the background therapy in addition to ENF could also contribute to the virologic response.
Response rates to TPV/r were also examined by TPV baseline phenotype. Based on data from the RESIST trials, baseline phenotype appears to be a predictor of response to TPV. Virologic response rates to TPV/r decreased when the shift in EC50 value from reference of TPV susceptibility was greater than three. In participants receiving TPV/r without concomitant ENF, the proportion of responders was 45% if TPV baseline susceptibility was three-fold or less, decreased to 21% responders when TPV susceptibility was between three and ten-fold, and dropped to 0% responders for a TPV baseline susceptibility change of greater than 10. Therefore, based on these results, a baseline TPV phenotype of less than three predicts the patient's HIV is susceptible to TPV. A baseline phenotype of three to ten predicts decreased susceptibility to TPV and greater than ten predicts resistance to TPV (Table 3).
The development of clinical resistance on TPV/r treatment was examined. The most common protease mutations that developed in isolates from treatment-experienced individuals who experienced virologic failure on TPV/r treatment were L33V/I/F, V82T and I84V. Notably, all available data came from treatment-experienced individuals and thus the resistance profile of TPV in treatment-naive individuals has not been characterized.
Following the complete review of the available data, the FDA in collaboration with the applicant must concisely describe the critical results of the submitted studies and include the most relevant and useful information for patients and clinicians in the package insert (drug label). In the TPV review, the multiple incremental breakpoints and subgroups were examined, but used the totality of the data to determine meaningful subgroups that provide useful information for clinicians and patients [7]. Rather than a single breakpoint for the number of baseline PI mutations or baseline phenotype, the preference is to use incremental subgroups representing maximal, reduced, or minimal response to treatment. For example, after an incremental approach examining response rates to TPV/r based on baseline TPV susceptibility, it was concluded that baseline TPV phenotype ranges 0-3, > 3-10, and > 10 best described maximal, reduced, and minimal response to TPV/r, respectively. Moreover, the response rates for the subgroups for maximal, reduced, or minimal response in the baseline phenotypic analysis, the number of baseline TPV-resistance-associated PI mutations, and the number of baseline PI mutations at L33, V82, I84, and/or L90 were comparable. In the package insert, the results of these analyses are shown in one table (Table 3). The consistent results across analyses support this approach.
In the Indications and Usage section of the TPV/r package insert, clinical resistance data was included to clarify the use of TPV/r. APTIVUS (tipranavir), co-administered with 200 mg of ritonavir, is indicated for combination antiretroviral treatment of HIV-1-infected adult patients with evidence of viral replication, who are highly treatment-experienced or have HIV-1 strains resistant to multiple protease inhibitors [14]. This indication is bulleted with additional information which stemmed from the evaluation of resistance in clinical studies.
* The use of other active agents with APTIVUS/ritonavir is associated with a greater likelihood of treatment response.
* Genotypic or phenotypic testing and/or treatment history should guide the use of APTIVUS/ritonavir. The number of baseline primary protease inhibitor mutations affects the virologic response to APTIVUS/ritonavir.
Genotypic and/or phenotypic analyses of baseline virus can aid in determining whether an antiretroviral drug has the potential to be an active component of a regimen before initiation of therapy in an HIV-infected patient. A comprehensive picture of drug resistance profiles is useful for clinicians when selecting antiretroviral drugs for patients with existing mutations in their HIV and reduced susceptibility to other antiretroviral drugs. Information on mutations that develop on treatment helps to assess the potential for resistance development to a drug and what future treatment options will remain after treatment with the drug. Therefore, this information is included in drug package inserts to assist prescribing physicians and HIV patients to choose optimal antiviral treatment regimens.
References
1. Food and Drug Administration. Guidance for Industry: Role of HIV Drug Resistance Testing in Antiretroviral Development. Made available 26 November 2004. Available at http://www.fda.gov/cder/guidance/5879dft.htm . Accessed: 23 October 2006.
[Context Link]
2. Poppe SM, Slade DE, Chong K, Hinshaw R, Pagano P, Markowitz M, et al. Antiviral activity of the dihydropyrone PNU-140690, a new nonpeptidic human immunodeficiency virus protease inhibitor. Antimicrob Agents Chemother 1997; 41:1058-1063.
[Context Link]
3. Larder B, Hertogs K, Bloor S, van den Eynde Ch, DeCian W, Wang Y, et al. Tipranavir inhibits broadly protease inhibitor-resistant HIV-1 clinical samples. AIDS 2000; 14:1943-1948.
[Context Link]
4. Rusconi S, La Seta Catamancio S, Citterio P, Kurtagic S, Violin M, Balotta C, et al. Susceptibility to PNU-140690 (Tipranavir) of human immunodeficiency virus type 1 isolates derived from subjects with multidrug resistance to other protease inhibitors. Antimicrob Agents Chemother 2000; 44:1328-1332.
[Context Link]
5. Doyon L, Tremblay S, Bourgon L, Wardrop E, Cordingley MG. Selection and characterization of HIV-1 showing reduced susceptibility to the non-peptidic protease inhibitor tipranavir. Antiviral Res 2005; 68:27-35.
[Context Link]
6. Food and Drug Administration Clinical and Statistical Reviews available at http://www.fda.gov/cder/foi/nda/2005/21814_000_AptivusTOC.htm Made available 22 June 2005. Accessed: 23 October 2006.
[Context Link]
7. Food and Drug Administration Microbiology Reviews available at http://www.fda.gov/cder/foi/nda/2005/21814_000_AptivusTOC.htm Made available 22 June 2005. Accessed: 23 October 2006.
[Context Link]
8. Hall D, McCallister S, Neubacher D, Kraft M and Mayers DL. Characterization of treatment-emergent resistance mutations in two phase II studies of tipranavir. XII International HIV Drug Resistance Workshop, June 2003, Cabo del Sol, Los Cabos, Mexico. Antiviral Therapy 8:S16.
[Context Link]
9. Hivinsite.ucsf.edu 2006.
[Context Link]
10. Johnson V, Brun-Vezinet F, Clotet B, Conway B, Kuritzkes D, Pillay D, et al. Update of drug resistance mutations in HIV-1:2005. Topics in HIV Medicine 2005; 13:51-57.
[Context Link]
11. Hivdb.stanford.edu 1998-2006.
[Context Link]
12. Kohlbrenner VM, Hall DB, Schapiro J, Baxter J, Boucher CA, Frank M, et al. Development of a tipranavir mutation score: analysis of protease mutations associated with phenotypic drug susceptibility and antiviral response in Phase II clinical trials. XIII International HIV Drug Resistance Workshop, June 2004, Tenerife, Canary Islands, Spain. Antiviral Therapy 9 (suppl):S143 [Abstract 129].
[Context Link]
13. Valdez H, Hall DB, Kohlbrenner VM, Boucher CA, Schapiro J, Baxter J, et al. Non-response to tipranavir is associated with pre-treatment resistance characterized by tipranavir phenotype or geneoytpic tipranavir score. XIV International HIV Drug Resistance Workshop, June 2005, Quebec City, Canada. Antiviral Therapy 10 (suppl 1):S29 [Abstract 27].
[Context Link]
14. Aptivus (Tipranavir) package insert. Boehringer Ingelheim, Ridgefield, Connecticut, USA.
[Context Link]
|
|
| |
| |
|
|
|