| |
ddI Clinical Cutoffs from the Jaguar Study
|
| |
| |
"Phenotypic Susceptibility to Didanosine Is Associated with Antiviral Activity in Treatment-Experienced Patients with HIV-1 Infection"
The Journal of Infectious Diseases Feb 1, 2007;195:392-398
Philippe Flandre,1 Colombe Chappey,5 Anne Genevieve Marcelin,2 Kirk Ryan,4 Jen-Fue Maa,4 Mike Bates,5 Daniel Seekins,4 Marie Charlotte Bernard,4 Vincent Calvez,2 and Jean Michel Molina3
1INSERM U720, 2Virology Hopital Pitie-Salpetriere, and 3Department of Infectious Diseases Hopital Saint-Louis, Paris, France; 4Bristol-Myers Squibb Virology, Plainsboro, New Jersey, and Rueil-Malmaison, France; 5Monogram Biosciences, Inc., South San Francisco, California
"....Our results demonstrate a relationship between baseline phenotypic susceptibility to ddI and antiviral activity of ddI at W4 in treatment-experienced patients with virologic failure who added ddI to their failing regimen..... The design of the JAGUAR study was appropriate for the investigation of such a relationship... the add-on design of the trial required that ddI be the only new drug introduced in the ART regimen, which ensured that any difference in virologic outcomes between the placebo and treatment arms could be reasonably ascribed to the addition of ddI.... "Clinical cutoffs" are critical in allowing the clinicians to properly interpret phenotypic resistance data and select active regimens for their patients..."
ABSTRACT
Objective. We investigated the relationship between human immunodeficiency virus (HIV) phenotypic susceptibility to didanosine and the antiviral activity of didanosine (ddI) in the JAGUAR study.
Methods. Baseline plasma HIV phenotypic susceptibility to ddI was assessed using a phenotype assay of patients randomized to receive ddI or placebo for 4 weeks in addition to their current regimen. Phenotypic susceptibility scores (PSSs) were then calculated for each sample. Associations between PSS and week 4 reductions in plasma HIV-1 RNA load or virologic response were assessed using linear regression and Jonckherre's test and the Wilcoxon and Cochran-Armitage tests, respectively.
Results.
In the ddI arm, a significant association between reduction in viral load and continuous PSS was observed (P < .0001). Using distinct categories, an increasing fold change (FC) in susceptibility to ddI was strongly associated with smaller reductions in plasma HIV-1 RNA load (P < .0001).
"....virologic response (defined as a reduction of 0.5 log10 in plasma HIV-1 RNA load at W4 or a W4 plasma HIV-1 RNA load <50 copies/mL)..."
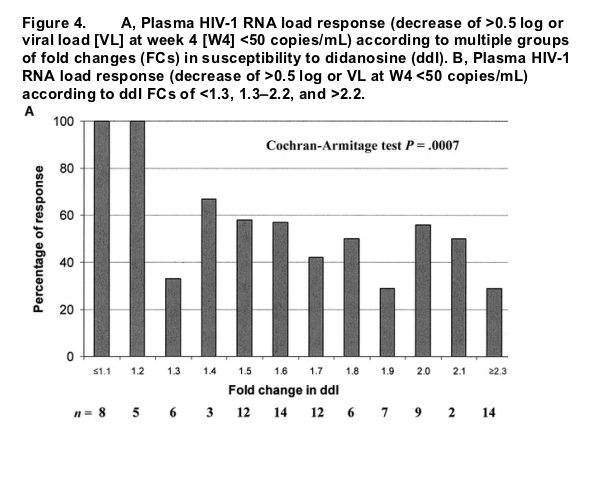
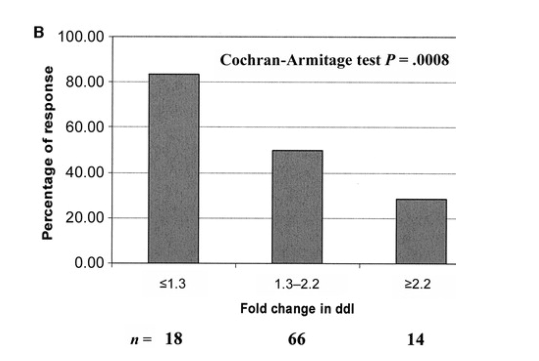
The proportion of virologic responders was 83% (15/18) for patients with a ddI FC <1.3, 50% (33/66) for patients with an FC of 1.3-2.2, and 29% (4/14) for patients with an FC >2.2 (P = .0008). After we determined these findings, 3 ddI FC categories were defined using 1.3 and 2.2 as thresholds.
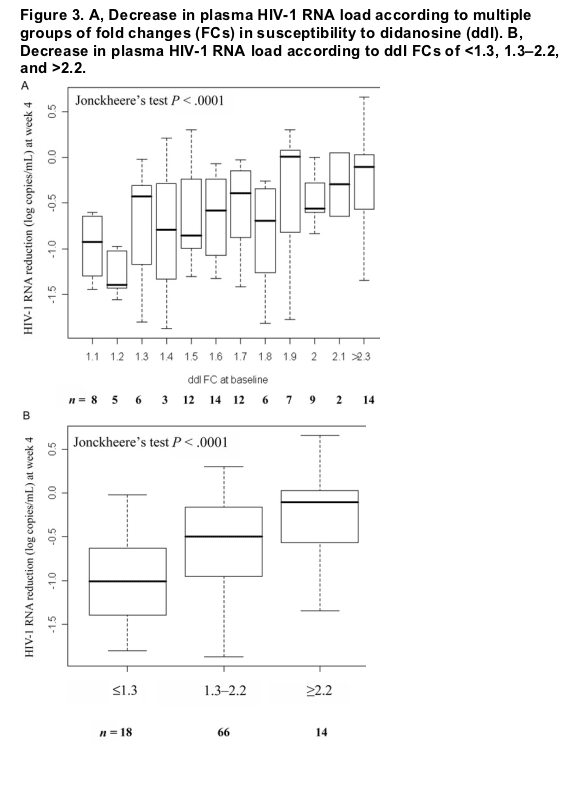
"....The association between increasing ddI FC categories and decreasing reductions in HIV-1 RNA load was found to be significant (P < .0001) when the Jonckheere test was used (figure 3A). The median decreases in log10 plasma HIV-1 RNA load were 1.01, 0.50, and 0.10 in patients with ddI FCs of <1.3, 2.2-1.3, and >2.2, respectively (figure 3B) (P < .0001, Jonckheere's test for trend)...."
Conclusions. The relationship between phenotypic susceptibility to ddI and reduction in plasma HIV-1 RNA load describes a continuum. The establishment of a lower clinical cutoff at 1.3 and an upper clinical cutoff at 2.2 are clinically relevant.
Background
Antiretroviral treatment (ART) may fail to achieve or maintain suppression of HIV replication because of drug resistance, lack of adherence, or suboptimal drug levels to >1 agent in a treatment regimen [1]. Although many drug combinations are potentially available, the number of those likely to achieve virological suppression is greatly limited by cross-resistance within drug classes. As a consequence, drug-resistance testing is now recommended for patients failing >1 prior ART regimen, to optimize their subsequent regimen [2, 3]. Genotypic resistance tests identify mutations in the viral genes targeted by antiviral drugs and have been used in several prospective studies to guide the choice of antiretroviral drugs [4-6]. The benefit of genotype testing is further improved by expert interpretation of the results, which may reveal complex patterns of mutations in patients who have had several ART regimens [7]. However, as multiple interpretation algorithms of genotypic tests are elaborated and as resistance to antiretroviral drugs evolves, these tests become difficult for clinicians to interpret. Phenotypic assays, on the other hand, have the potential advantage of providing a quantitative assessment of viral susceptibility to all antiviral drugs and, thus, of measuring the continuum of susceptibility that may exist between virus and drug. Although a prospective trial of phenotypic resistance testing did not demonstrate any benefit of treatment decisions based on phenotypic guidance for the test cohort as a whole, patients with protease-inhibitor resistance or with greater treatment experience at baseline did appear to benefit [8]. Of note, the California Collaborative Treatment Group (CCTG) 575 study demonstrated the importance of accurate clinical break points in the interpretation of phenotypic resistance data, because the failure of phenotypic testing to clearly demonstrate an advantage was at least partially attributed to inaccuracy in the clinical cutoffs for multiple drugs [8]. The JAGUAR study, because of its design and carefully collected clinical outcome data, presented an excellent opportunity for the identification of accurate phenotypic cutoffs for didanosine (ddI).
In the present study, we used data from the recently completed JAGUAR study [9] to investigate the ability of baseline phenotypic susceptibility to ddI to predict virologic outcome. In this add-on, randomized, placebo-controlled trial, ddI was the only drug added to a failing regimen for the assessment of its short-term antiviral activity and safety. The primary objective of the analysis was to investigate the relationship between phenotypic susceptibility to ddI at baseline and virologic response at 4 weeks, using the PhenoSense assay (Monogram Biosciences). A second objective was to identify and refine clinically relevant break points associated with reduced susceptibility to ddI in vivo.
MATERIALS AND METHODS
Study design. The JAGUAR study was a multicenter, add-on, randomized, double-blind, placebo-controlled, comparative trial performed at 29 sites in France [9]. Patients were randomly assigned in a 2 : 1 ratio to add ddI (n = 111) or a matching placebo (n = 57) to their current antiretroviral regimen for 4 weeks. ddI was provided as 1 enteric-coated capsule (400 mg for patients weighing 60 kg and 250 mg for patients weighing <60 kg) and placebo as 1 matching capsule. At 4 weeks, there was a significant decrease in mean plasma HIV-1 RNA load of -0.56 log10 copies/mL in patients who received ddI, compared with an increase of 0.07 log10 copies/mL in those who received the placebo (P < .001) [9].
Plasma HIV-1 RNA measurements and end points. Plasma HIV-1 RNA loads were measured in a central laboratory using the Amplicor HIV-1 Monitor kit (Cobas 1.5 test; Roche Diagnostic Systems) at baseline, week 2, and week 4 (W4). The lower limit of quantification was 50 copies/mL. Two virologic outcomes were considered: the reduction in log10 plasma HIV-1 RNA load at W4 (continuous variable) and a virologic response defined as the proportion of patients with a reduction of >0.5 log10 in plasma HIV-1 RNA load at W4 or with a W4 plasma HIV-1 RNA load <50 copies/mL (dichotomous variable).
Phenotypic resistance assay to ddI. The phenotypic measures of viral susceptibility to ddI in patient plasma samples were determined using the PhenoSense HIV assay (Monogram Biosciences) [10]. Phenotypic susceptibility results were expressed as the fold change (FC) in susceptibility to ddI and were calculated as the ratio of the IC50 of patient-derived virus to the IC50 of the drug-susceptible control virus NL4-3. Reproducibility data were obtained from 20 repeated phenotype results for 3 site-directed mutants: WAC 2 (GenBank accession number DQ988164), WAC 8 (DQ988165), and WAC 10 (DQ988166). These 3 mutants are clonal viruses derived from a clinical plasma sample that have distinct patterns of drug-resistance mutations. The WAC virus genotypes were defined by the amino acids that differed from those of the wild-type virus NL4-3. The WAC 2 virus genotype was 10I, 35D, 36I, 54V, 62V, 63P, 71V, 82T, 84V, and 90M in protease and 35I, 39A, 41L, 67N, 102K, 103N, 122E, 158S, 162S, 184V, 200A, 203D, 210W, 211K, 215Y, 218E, 219Q, 272P, 288T, and 297R in reverse transcriptase. The WAC 8 virus genotype was 10I, 13V, 20I, 33I, 35D, 36I, 37D, 54V, 58E, 63P, 66F, 71V, 82T, and 90M in protease and 13E, 35M, 67N, 69D, 70R, 102K, 108I, 109V, 121Y, 122E, 162S, 181C, 189L, 200A, 202T, 208Y, 211K, 215F, 218E, 219Q, 221Y, 227L, 228H, 277K, 283I, 284K, 286A, 293I, and 297K in reverse transcriptase. The WAC 10 virus genotype was 10F, 14R, 15V, 30N, 37D, 41K, 63P, 77I, and 88D in protease and 35T, 39A, 41L, 60I, 67N, 68G, 70R, 90I, 102K, 103N, 135T, 162Y, 184V, 200A, 215Y, 219E, 277K, 293I, and 294Q in reverse transcriptase. The mean FC and coefficient of variation (CV) for each mutant were calculated.
Phenotypic susceptibility scores (PSSs). Three PSSs were assessed. For each score, the FC ranged from 0 (ddI-resistant virus) to 1 (ddI-susceptible virus). A partially continuous PSS (PSS1) was calculated as follows: the value 0 was assigned to observations with FC >10; 1 to FC values less than or equal to the lower cutoff (LCO); and a value between 0 and 1, calculated as 1 - (FC - LCO)/(10 - LCO), for observations with an FC between the LCO and 10, as described elsewhere [11]. Distinct LCOs were investigated. A dichotomous PSS (PSS2) assigned 1 to observations with a ddI FC less than or equal to the LCO and 0 otherwise. A completely continuous PSS (PSS3) was designed to reflect the distribution of the observed FCs in our data. PSS3 was calculated as 1 - {log10(FC) - log10[MIN(FC)] / (log10[MAX(FC)] - log10[MIN(FC)]}, where MIN(FC) and MAX(FC) correspond to the minimum and maximum observed value of ddI FCs in the data set (after the exclusion of an outlier with a very high FC value of 22). The log10 transformation of the ddI FC was used because preliminary results indicated a better linear fit.
Statistical methods. The linear regression model was used to evaluate the association between reductions in plasma HIV-1 RNA load at W4 and ddI FC, both used as a continuous variables, and the Wilcoxon test was used for testing the association between reduction in plasma HIV-1 RNA load and dichotomous ddI FC (as defined in PSS2). Multiple categories of ddI FC were examined, and the trend toward decreasing reductions in HIV-1 RNA load across FC categories was tested using the Jonckheere test [12]. The test for trend in virologic response (dichotomous variable) across FC categories used the Cochran-Armitage test. Distinct couples of cutoffs were explored, and the choice of the final cutoffs was based on both P values and practical relevance.
RESULTS
PhenoSense assay reproducibility for ddI. Figure 1 displays the results of 20 replicate measures of FC ddI for each of the 3 mutants. For the WAC 8 and WAC 10 mutants, all measures stayed within a ddI FC of ±0.29 and ±0.49, respectively. For the WAC 2 mutant, the range covered a ddI FC of ±1.01. The CVs were 8.2%, 8.6%, and 12% for WAC 8, WAC 10, and WAC 2, respectively, which indicated that the assay is highly reproducible. With the WACs, we assessed the assay's reproducibility on clones. Mean ddI FC IC50 values obtained for 20 clones were 0.84, 1.63, and 1.81 for WAC 8, WAC 10, and WAC 2, respectively, and the 95% confidence intervals were 0.82-0.89, 1.56-1.69, and 1.71-1.91. In a previous publication, the natural variation in drug susceptibility of nearly 3000 clinical isolates defined as wild-type viruses for all drugs was described [13]. For ddI, the median FC IC50 value for wild-type viruses was found to be 0.9, with the 99th percentile being 1.3, which corresponds to the biological cutoff. A narrow confidence interval around the median for wild-type viruses demonstrates the ability of the PhenoSense HIV assay to reliably measure subtle but clinically relevant changes in drug susceptibility. This precision of assay allows assessing with confidence that mutational alteration leading to a slight increase in phenotypic resistance to ddI would be detected at an FC of >1.3.
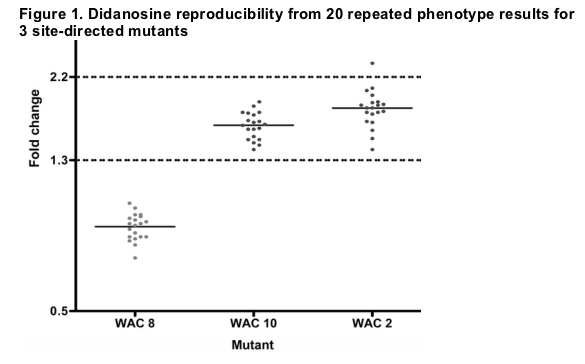
Phenotypic susceptibility to ddI in the JAGUAR study. ddI FCs at baseline and virologic outcomes at W4 were obtained for 146 patients: 98 in the ddI group and 48 in the placebo group. Median ddI FCs (minimum, maximum [interquartile range]) were 1.65 (0.86, 22 [1.46-1.97]) and 1.63 (1.03, 2.9 [1.32-2.08]) in the ddI and placebo groups, respectively. The relationship between reductions in log10 plasma HIV-1 RNA load at W4 and ddI FC at baseline for the 2 groups is depicted in figure 2. The patient with a ddI FC of 22 was considered to be an outlier and had baseline mutations associated with multidrug resistance (A62V, K65R, V75I, F77L Y115F, F116Y, Q151M, and M184V).
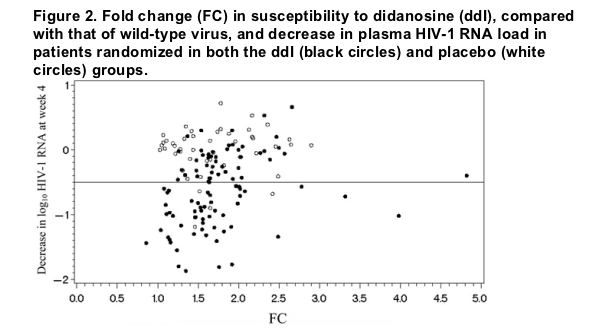
Reduction in plasma HIV-1 RNA load at W4 as a virologic outcome. A number of univariate linear models were fitted to the data using the reduction in plasma HIV-1 RNA load as a virologic outcome. In the ddI group, ddI FC and PSS1 were not predictive of a reduction in viral load, whereas log-transformed ddI FC and PSS3 were highly predictive (P = .004 and P < .0001, respectively). Although both log10 ddI FC and PSS3 were significantly associated with a reduction in plasma HIV-1 RNA load at W4, they explained only 12% and 8% of the reduction, respectively (table 1). The variability explained may appear to be modest, but fitting a linear regression with baseline plasma HIV-1 RNA load as a single variable resulted in P = .0047 (R2 = 0.0802), and regression with baseline CD4 cell count resulted in P = .0086 (R2 = 0.0735). When the dichotomous PSS2 score was used, median reductions in log10-transformed plasma HIV-1 RNA load were 0.96 and 0.43 copies/mL in patients defined as susceptible (PSS2, 1) and resistant (PSS2, 0), respectively (P = .001, Wilcoxon test). None of these variables were significantly associated with a reduction in plasma HIV-1 RNA load in patients from the placebo group.

The association between increasing ddI FC categories and decreasing reductions in HIV-1 RNA load was found to be significant (P < .0001) when the Jonckheere test was used (figure 3A). The median decreases in log10 plasma HIV-1 RNA load were 1.01, 0.50, and 0.10 in patients with ddI FCs of <1.3, 2.2-1.3, and >2.2, respectively (figure 3B) (P < .0001, Jonckheere's test for trend). Similar P values were also obtained with 3 other pairs of cutoffs-1.3/2.0, 1.6/2.0, and 1.6/2.2- but the intermediate FC interval was even more narrow than that of 1.3/2.2, which limited the use of such intervals in practice. In addition, the threshold of 1.3 is also the biological cutoff for ddI. Similar analysis applied to the placebo group did not demonstrate any significant association between ddI FC categories and reductions in plasma HIV-1 RNA load (data not shown).
Virologic response at W4 as a virologic outcome. Nine (9%) of 98 patients reached an HIV-1 RNA load <50 copies/mL at W4. In the ddI group, the proportion of patients with a virologic response (defined as a reduction of >0.5 log10 in plasma HIV-1 RNA load at W4 or a W4 plasma HIV-1 RNA load <50 copies/mL) by ddI FC categories varied from 100% to 29%, with large variability in sample size for individual FC categories (figure 4A). The trend was statistically significant, which indicates a higher percentage of virologic response in the categories with lower ddI FCs (P = .0007, Cochran-Armitage test). The proportion of responders was 83% (15/18 patients) with a ddI FC <1.3, 50% (33/66 patients) with FC 1.3-2.2, and 29% (4/14 patients) with >2.2 (P = .0008, Cochran-Armitage test). In accordance with these findings, 3 categories were defined using 1.3 and 2.2 as thresholds for both reduction in plasma HIV-1 RNA load and virologic response (figure 4B). Similar analyses performed in the placebo group did not show a significant association between virologic response and ddI FC categories (data not shown).
DISCUSSION
Our results demonstrate a relationship between baseline phenotypic susceptibility to ddI and antiviral activity of ddI at W4 in treatment-experienced patients with virologic failure who added ddI to their failing regimen. The design of the JAGUAR study was appropriate for the investigation of such a relationship for at least 2 reasons. First, the add-on design of the trial required that ddI be the only new drug introduced in the ART regimen, which ensured that any difference in virologic outcomes between the placebo and treatment arms could be reasonably ascribed to the addition of ddI. Second, a continuous outcome such as the reduction in plasma HIV-1 RNA load is more informative than a binary virologic response, especially when one is searching for a continuous relationship. In addition, assessing reductions in plasma HIV-1 RNA load at W4 led to a low proportion of patients being censored by the limit of detection of the assay, which allowed the use of standard statistical methods instead of methods specific to censored observations [14, 15].
In clinical practice, it is usually helpful to classify the susceptibility of a pathogen to drugs into at least 3 categories (high, low, or intermediate) to determine whether the use of a drug is appropriate. In the field of HIV infection, however, the use of phenotypic susceptibility assays has been limited by an insufficient understanding of the relationship between FCs in IC50 values and the probability of virologic response in any given patient. "Clinical cutoffs" are critical in allowing the clinicians to properly interpret phenotypic resistance data and select active regimens for their patients. The CCTG 575 study, which showed no benefit to phenotypically guided regimen selection, used an arbitrary clinical cutoff of <2.5 for all drugs, including ddI, so that viruses with a measured ddI FC of 2.5 were uniformly called "sensitive." It was argued that the lack of benefit of the phenotypic arm in CCTG 575 could be partially explained by inaccurate clinical cutoffs for a number of drugs, particularly nucleoside analogues, which resulted in suboptimal ART selection [8]. Recent data have suggested that a cutoff of 2.5 may be too high for ddI and stavudine and too low for abacavir [16, 17]. On the basis of the results of the present study, we now know that the probability of virologic response to ddI begins to wane at FC values of 1.3 and decreases to <30% by the time the FC increases to 2.2, let alone 2.5. Similar analyses of abacavir add-on studies have established a lower clinical cutoff of 4.5 for that drug. Examination of the drug usage patterns in CCTG 575 showed a disproportionately high frequency of use of ddI and a correspondingly low frequency of use of abacavir in the phenotypically guided arm of that study, compared with that in the control arm, exactly as one would predict if the cutoffs for these drugs were erroneously high or low, respectively [8].
We classified phenotypic susceptibility to ddI into 3 different categories, which were associated with distinct levels of reductions in plasma HIV-1 RNA load and proportions of virologic responses. The use of phenotypic FC cutoffs of 1.3 and 2.2 allowed us to identify patients with a high (84%), intermediate (50%), or low (29%) probability of virologic response, defined as a reduction in viral load of at least 0.5 log10 HIV-1 RNA or a level of HIV-1 RNA <50 copies/mL (figure 4B). The same cutoffs were associated with a median decrease in log10 plasma HIV-1 RNA load at W4 of 1.01, 0.5, and 0.1, respectively (figure 3B). The ddI cutoff used in the PhenoSense assay was 1.7 before the results of our study (available at: http://www.monogramhiv.com/assays/hcp/phenosenseHIV.aspx). Using this 1.7 cutoff would have led us to obtain only 62% of virologic responders among patients with phenotypic susceptibility to ddI and up to 42% of virologic responders among patients with phenotypic resistance to ddI (data not shown). This finding emphasizes the importance of defining appropriate clinical cutoffs for phenotypic testing and offers further support to the notion that the use of incorrect clinical cutoffs may explain the results of CCTG 575. Thus, in accordance with the results of our study, the lower clinical cutoff for ddI was refined from 1.7 to 1.3. The precision and reproducibility of the PhenoSense assay for ddI (figure 1) allowed this decrease of the clinical cutoff value.
However, our study does have some limitations. Our results were obtained using the PhenoSense assay and cannot be extended to other phenotypic assays because of differences in methodology and performance characteristics. Efforts should be made to pursue the rigorous definition of clinical cutoffs for all phenotypic assays, because there is now evidence to suggest that the accuracy of clinical cutoffs can have a marked effect on the ability of any assay to predict outcome and, thus, to properly offer the best chance of choosing an effective regimen for the patient. In addition, the population of patients in whom the study was performed may not be representative of all treatment-experienced patients.
Although we believe that the results we obtained in the present study are valid and that the design of the clinical trial and the statistical tests were appropriate to study and define clinically relevant cutoffs of phenotypic assays [9], efforts should be made to validate the results of these analyses using additional, larger data sets. This raises methodological challenges, because patients are rarely if ever treated in a way that would facilitate the easy analysis of cause and effect, as in add-on studies like JAGUAR. However, as larger databases are created and more information becomes available regarding the clinically relevant break points for each drug in the HIV armamentarium, it may become possible to account for the confounding influence on virologic response of all but the drug under study and, thus, to examine the FC-response relationship in a "real world" setting. Such approaches may ultimately lead to more accurate assessments of antiviral activity for individual patients, the selection of more effective regimens resulting in more durable responses, and less drug resistance.
References
1. Hirsch MS, Brun-Vezinet F, D'Aquila RT, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA 2000; 283:2417-26. First citation in article | PubMed | CrossRef
2. US Department of Health and Human Services Panel on Clinical Practices for Treatment of HIV Infection. Guidelines for the use of antiretroviral agents in HIV-1 infected adults and adolescents. Available at: http://www.aidsinfo.nih.gov. Accessed 10 November 2003. First citation in article
3. European guidelines from the clinical management and treatment of HIV-infected adults in Europe. AIDS 2003; 17(Suppl 2):S3-26. First citation in article | PubMed | CrossRef
4. Durant J, Clevenbergh P, Halfon P, et al. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 1999; 353:2195-9. First citation in article | PubMed | CrossRef
5. Meynard JL, Vray M, Morand-Joubert L, et al. Phenotypic or genotypic resistance testing for choosing antiretroviral therapy after treatment failure: a randomised trial. AIDS 2002; 16:727-36. First citation in article | PubMed | CrossRef
6. Baxter JD, Mayers DL, Wentworth DN, et al. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. AIDS 2000; 14:F83-93. First citation in article | PubMed | CrossRef
7. Tural C, Ruiz L, Holtzer C, et al. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS 2002; 16:209-18. First citation in article | PubMed | CrossRef
8. Haubrich RH, Kemper CA, Hellmann NS, et al. A randomized prospective study of phenotype susceptibility testing versus standard of care to manage antiretroviral therapy: CCTG 575. AIDS 2005; 19:295-302. First citation in article | PubMed
9. Molina J-M, Marcelin AG, Pavie J, et al. Didanosine in HIV-1-infected patients experiencing failure of antiretroviral therapy: a randomized placebo-controlled trial. J Infect Dis 2005; 191:840-7. First citation in article | Full Text | PubMed
10. Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother 2000; 44:920-8. First citation in article | PubMed | CrossRef
11. Katzenstein DA, Bosch RJ, Hellmann NS, et al. Phenotypic susceptibility and virological outcome in nucleoside-experienced patients receiving three or four antiretroviral drugs. AIDS 2003; 17:821-30. First citation in article | PubMed | CrossRef
12. Lehmann EL. Nonparametrics: statistical methods based on ranks. San Francisco: Holden-Day, 1975. First citation in article
13. Parkin NT, Hellmann NS, Whitcomb JM, Kiss L, Chappey C, Petropoulos CJ. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob Agents Chemother 2004; 48:437-43. First citation in article | PubMed | CrossRef
14. Flandre P, Durier C, Descamps D, Launay O, Joly V. On the use of magnitude of reduction in HIV-1 RNA in clinical trials: statistical analysis and potential biases. J Acquir Immune Defic Syndr 2002; 30:59-64. First citation in article | PubMed | CrossRef
15. Flandre P, Alcais A, Descamps D, Morand-Joubert L, Joly V. Estimating and comparing reduction in HIV-1 RNA in clinical trials using methods for interval censored data. J Acquir Immune Defic Syndr 2004; 35:286-92. First citation in article | PubMed
16. Lanier E, Hellmann N, Scott J, et al. Determination of a clinically relevant phenotypic resistance cutoff for abacavir using the Phenosense assay [abstract 254]. In: Program and abstracts of the 8th Conference on Retroviruses and Opportunistic Infections (Chicago). Alexandria, VA: Foundation for Retrovirology and Human Health, 2001. First citation in article
17. Kempf D, Brun S, Rode R, et al. Identification of clinically relevant phenotypic and genotypic breakpoints for ABT-378r in multiple PI-experienced, NNRTI-naive patients. Antivir Ther 2000; 5(Suppl 3):70. First citation in article
|
|
| |
| |
|
|
|