| |
Prognostic Value of HIV-1 RNA, CD4 Cell Count, and CD4 Cell Count Slope for Progression to AIDS and Death in Untreated HIV-1 Infection
|
| |
| |
JAMA
June 6, 2007
Research Letter
To the Editor: In a study reporting that plasma human immunodeficiency virus 1 (HIV-1) RNA explains less than 10% of the variability in CD4 cell count slope in patients with untreated HIV-1 infection, Rodriguez et al1 questioned viral replication as the main determinant of progressive immunodeficiency. That study did not include the clinically important outcomes of AIDS or death and did not provide data on variability of CD4 cell count slopes. We therefore evaluated the prognostic strength of HIV-1 RNA, CD4 cell count,2-3 and CD4 cell count slope for clinical outcomes, as well as the variance of CD4 cell count slope.
Methods
The study population comprised 1640 HIV-seropositive participants in the Multicenter AIDS Cohort Study (MACS).4 Baseline was the earliest semiannual visit after the first seropositive visit at which plasma HIV-1 RNA and CD4 cell count were available. Measurement of HIV-1 RNA was obtained by reverse transcriptase-polymerase chain reaction (RT-PCR; Amplicor HIV Monitor Assay, Roche Diagnostics, Nutley, NJ) or bDNA (Chiron Corp, Emeryville, Calif), with bDNA results converted to RT-PCR values.3
Values of HIV-1 RNA that were below detection limits (n = 77) were imputed using parametric methods for left-censored data.5 Individuals' CD4 cell count slopes and coefficients of variation (CV; standard error/|slope|) were determined by linear regressions between baseline and mid-1988 (with 1518 [93%] of the participants having at least 3 data points), comparable with the 2- to 3-year interval analyzed by Rodriguez et al.1 Models with random intercepts and slopes for longitudinal CD4 cell counts were used to assess serial correlation and to provide Bayes estimates of slopes.6 Coefficients of determination (R2) for censored survival data, derived from generalized gamma regressions to include censored observations,7 were used to quantify percentage of variability explained by predictors for log-scaled time to AIDS (1993 definition, not including CD4 cell counts <200/μL), CD4 count of less than 200/μL (n = 1472 without CD4 cell counts <200/μL at or prior to baseline), and death. Follow-up was censored in December 1990, before widespread use of combination nucleoside reverse transcriptase inhibitor therapy, which would influence outcome. Statistical significance was defined by 2-sided P<.05. Confidence intervals were based on 100 bootstrap samples. Statistical analyses were performed with SAS software, version 9.1 (SAS Institute Inc, Cary, NC) and S-PLUS software, version 7.0 (Insightful, Seattle, Wash).
Results
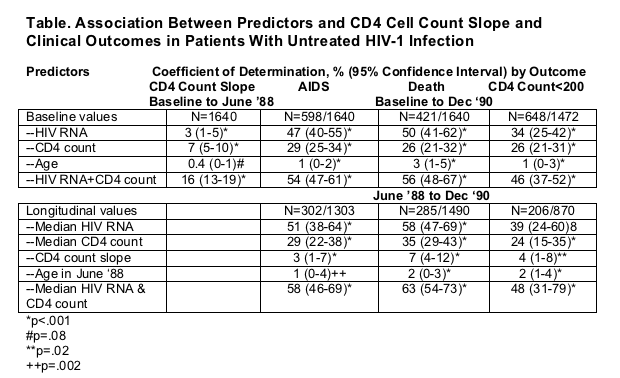
Median baseline values were as follows: HIV-1 RNA, 22 002 (interquartile range [IQR], 7957-60 414) copies/mL; CD4 cell count, 544/μL (IQR, 382-735/μL); age, 34 years (IQR, 29-38 years); and CD4 cell count slope, -64/μL per year (IQR, -136 to -8/μL per year). By December 1990, 598 (37%) of 1640 participants had developed AIDS, 648 (44%) of 1472 had reached a CD4 count of less than 200/μL, and 421 (26%) of 1640 had died (Table).
Baseline HIV-1 RNA explained 3% of the variability in CD4 cell count slope (Table). CD4 cell count and age explained 7% and less than 1% of the variability, respectively. Baseline HIV-1 RNA measurement explained 47% and 50% of the variability in times to AIDS and death, respectively. Baseline CD4 cell count explained 29% and 26% of the variability in times to AIDS and death, respectively; age explained 1% and 3%, respectively. HIV-1 RNA and CD4 cell count explained 34% and 26% of the variability in time to CD4 count of less than 200/μL, respectively.
Using longitudinal variable data obtained prior to mid-1988 to predict clinical outcomes observed between mid-1988 and December 1990, CD4 cell count slope explained 3% and 7% of the times to AIDS and death, respectively (Table). Median HIV-1 RNA explained 51% and 58% of the variability in AIDS and death, and median CD4 cell count explained 29% and 35% of the variability in AIDS and death, respectively.
The median CV of CD4 cell count slopes was 68% (IQR, 35%-167%), with 37% of the slopes having a CV of more than 100%. Random regression models showed r = 0.009 among individuals' serial CD4 cell counts. The median CV of empirical Bayes estimates of slopes (shrunk toward population averages) was 70%, with 1 of 6 of these slopes having a CV of more than 100%.
COMMENT
In patients with untreated HIV infection, a single HIV-1 RNA measurement was the strongest baseline predictor of times to AIDS and death, explaining about half of the variability in these clinically important outcomes.
Consistent with Rodriguez et al,1 baseline HIV-1 RNA explained only 3% of the variability in CD4 cell count slope. The large variance of CD4 cell count slopes may be the reason for this finding and for the small amount of variability in times to AIDS and death explained by CD4 cell count slope. The lower coefficient of determination of HIV-1 RNA for time to CD4 cell count of less than 200/μL compared with AIDS or death is also consistent with the variability of CD4 cell count and slope.
The prognostic strength of HIV-1 RNA is consistent with a central role of viral replication, manifest as viremia, in AIDS pathogenesis. It supports the use of HIV-1 RNA for estimating prognosis in untreated HIV-1 infection.
Financial Disclosures: Dr Mellors reports that he is or has been a consultant to Abbott Laboratories, Bristol-Myers Squibb, Agouron Pharmaceuticals, Boehringer-Ingelheim, Gilead Sciences, GlaxoSmithKline, Intelligent Therapeutic Solutions, Merck, Noviro/Idenix, Pfizer, Pharmasset, Trimeris, and Visible Genetics; has owned or currently owns stock or stock options in Achillion Pharmaceuticals, Noviro/Idenix, Intelligent Therapeutic Solutions, Pharmasset, Triangle Pharmaceuticals, and Virco-Tibotec; and has filed the following patents: US Patent Application No. 60/646 593, 2005-methods for high-efficiency single-genome sequencing of HIV; US Patent Application No. PCT/US07/02369-HIV-1 mutations at codon 371 and 509 of reverse transcriptase increase resistance to nucleoside analogs such as 3'-azidothymidine; and US Patent Application No. 60/813 068-multigenome sequencing methods. No other financial disclosures were reported.
Funding/Support: MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute and the National Heart, Lung, and Blood Institute.
Role of the Sponsors: The sponsors had no role in the design and conduct of the study; in the collection, management, analysis, and interpretation of the data; or in the preparation of the manuscript.
Editors' Note: While JAMA generally requires that study data be no more than 3 to 4 years old, the editors concluded that this study question could be addressed only by a database collected before the use of combination antiretroviral therapy.
John W. Mellors, MD
mellors@dom.pitt.edu
University of Pittsburgh
Pittsburgh, Pa
Joseph B. Margolick, MD, PhD
Johns Hopkins University
Baltimore, Md
John P. Phair, MD
Northwestern University
Chicago, Ill
Charles R. Rinaldo, PhD
University of Pittsburgh
Pittsburgh, Pa
Roger Detels, MD, MS
University of California, Los Angeles
Lisa P. Jacobson, ScD; Alvaro Munoz, PhD
Johns Hopkins University
Baltimore, Md
1. Rodriguez B, Sethi AK, Cheruvu VK, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. JAMA. 2006;296:1498-1506. FREE FULL TEXT
2. Mellors JW, Rinaldo CR, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167-1170. ABSTRACT
3. Mellors JW, Munoz A, Giorgi JV, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946-954. FREE FULL TEXT
4. Kaslow RA, Ostrow DG, Detels R, et al. The Multicenter AIDS Cohort Study: rationale, organization and selected characteristics of the participants. Am J Epidemiol. 1987;126:310-318. ISI | PUBMED
5. Lau B, Gange SJ. Methods for the analysis of continuous biomarker assay data with increased sensitivity. Epidemiology. 2004;15:724-732. FULL TEXT | ISI | PUBMED
6. Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963-974. FULL TEXT | ISI | PUBMED
7. Cox C, Chu H, Schneider M, Munoz A. Parametric survival analysis and taxonomy of hazard functions for the generalized gamma distribution [published online ahead of print March 6, 2007]. Stat Med. doi:10.1002/sim2836.
|
|
| |
| |
|
|
|