 |
 |
 |
| |
Online Coreceptor Algorithm Predictors Won't Work in Clinic
|
| |
| |
Mark Mascolini
Free online algorithms that figure which CD4-cell coreceptor HIV uses aren't ready to replace the slower, expensive Trofile phenotype devised by Monogram Biosciences [1]. Viviane Lima (BC Centre for Excellence in HIV/AIDS, Vancouver) and coworkers reached that conclusion after comparing coreceptor preference ("tropism") calls by Trofile versus six online tools.
This analysis involved 953 viral samples that underwent both V3 loop sequencing and Trofile phenotyping. (Viruses using different coreceptors differ in the V3 region of the HIV's envelope gene sequence.) Lima ran the sequences through six algorithms built to determine coreceptor tropism:
- Positive charges at V3 positions 11 and/or 25 (the "11/25" rule)
- Neural Network
- Position-Specific Scoring Matrix trained on the syncytium-inducing phenotype (PSSM SI/NSI)
- PSSM trained on the X4 or R5 coreceptor phenotype (PSSM X4/R5)
- Support Vector Machine (SVM) genomaniac
- SVM geno2pheno
Specificity of calling coreceptor use was high for all algorithms, but sensitivity was abysmal (Table). (Sensitivity is the probability that a condition, such as CXCR4 tropism, is present; specificity is the probability that a condition is not present [2].) Even when Lima adjusted this analysis by increasing the aggressiveness of calling viral samples CXCR4-capable, sensitivities improved only into a range of 24% to 50%.
Sensitivity and specificity of genotype algorithms to call HIV coreceptor use
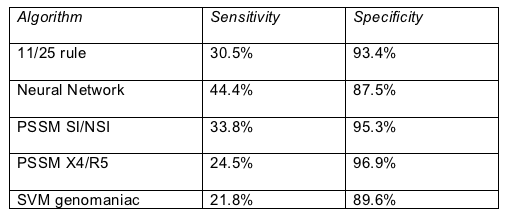
Lima cloned amplified polymerase chain reaction products to improve detection of low-level CXCR4 variants. But when the CXCR4-using part of a viral population dropped below about 23%, detection of these minority viruses in a bulk genotype became unreliable. She also found that as a person's CD4 count fell, the positive predictive value of all methods increased significantly (P < 0.05). In other words, all methods were more likely to be accurate in people with lower CD4 counts.
Why did the algorithms fare so poorly? Lima suggested three reasons:
1. The algorithms were "trained" on data from viral clones, but she tested them on clinically derived data (as they would be if used by clinicians).
2. The algorithms were "trained" on an eclectic mix of phenotyping assays.
3. Bulk sequencing--the source of genotypes in this analysis--misses minority viral species.
Lima suggested five ways to improve algorithms--tactics primarily of interest to experts in this field. She concluded that, for now, these genotype-based tools are inadequate for reckoning tropism of clinical viral samples.
Charles Boucher (University Medical Centre, Utrecht) disagreed, arguing that the algorithms' high specificity is clinically useful. A 90% specificity, for example, means that an algorithm will tell you correctly nine out of 10 times whether a viral sample is not CXCR4 tropic. If you can conclude on that basis that a certain virus uses CCR5, you can be reasonably sure that a CCR5 antagonist will work.
But not everyone agreed with that approach. Douglas Richman (University of California, San Diego) wondered if 90% specificity is good enough. And Richard Harrigan, a coauthor of this study, said it would be better to fix the low sensitivity than to rely on the 90% specificity.
References
1. Low A, Chan D, Dong W, et al. Current implementations of HIV genotyping
algorithms are inadequate for the prediction of X4 coreceptor usage in clinical isolates from population-based V3 sequences: approaches to improvement. Antiviral Therapy. 2007;12:S164. Abstract 149.
2. Sensitivity, Specificity and Predictive Values. http://research.med.umkc.edu/tlwbiostats/sens_specif_predval.html.
|
| |
|
 |
 |
|
|