| |
Long-term follow-up of successful hepatitis C virus therapy: waning immune responses and disappearance of liver disease are consistent with cure - HCV traces in PBMC of 3/54 SVRs disappear after 9 yrs
|
| |
| |
Download the PDF here
Alimentary Pharmacology & Therapeutics March 2015
"Patients with SVR after treatment of chronic hepatitis C with interferon or peginterferon alpha-2b ± ribavirin were identified in the participating clinics patient records.....had achieved SVR at least 5 years prior to inclusion....liver biochemistry, histology and elastography were evaluated. Liver biopsies, plasma and peripheral blood mononuclear cells (PBMCs) were tested for minute amounts of HCV RNA. HCV-specific T-cell responses were monitored by ELISpot and pentamer staining, and humoral responses by measuring HCV nonstructural (NS)3-specific antibodies and virus neutralization.......median follow-up time was 9.8 years (range 5-20) post-SVR, and the median duration of infection prior to SVR was 18.4 years.....All patients were negative for HCV RNA in serum at follow-up using the routine COBAS AmpliPrep/COBAS TaqMan HCV test.....
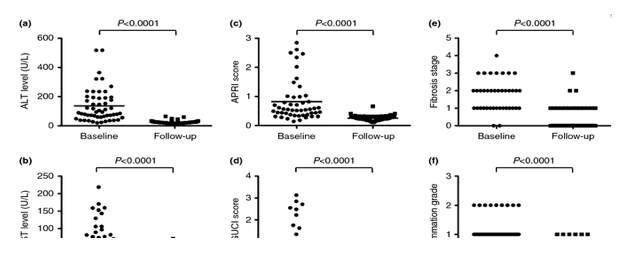
......Clinical, biochemical and histological parameters indicating liver disease were compared between samples taken at baseline (before the start of treatment) and at follow-up (Figure 1). Both the ALT (from 136.78 to 24.14 U/L) and AST levels (from 66.66 to 25.14 U/L) in the serum decreased significantly (P < 0.0001, Mann-Whitney U-test) from baseline to follow-up (Figure 1a,b and Table 1). Similarly, we detected a significant decrease (P < 0.0001, Mann-Whitney U-test) of the APRI (from 0.82 to 0.26) and GUCI scores (from 0.89 to 0.27) from baseline to follow-up (Figure 1c,d and Table 1). From 39/54 patients (72%), liver biopsies were available both at baseline and at follow-up and were used to determine the inflammation grade and the fibrosis stage. Both decreased significantly (P < 0.0001, Mann-Whitney U-test) from baseline to follow-up (Figure 1e,f and Table 1). The mean FibroScan elasticity level at follow-up was 4.9 kPa (range 2.4-8.8 kPa) confirming the histological analysis (Table 1). Thus, 5-20 years after SVR, the degree of liver disease had regressed and in the majority normalised according to all measured clinical, biological and histological markers....
............Plasma (all 54 patients), PBMCs (all 54 patients) and liver biopsies (40/54 patients) from treatment-recovered patients were tested for HCV RNA in a combined ultracentrifugation/RT-PCR method with a high sensitivity of 2 copies/mL (Figure 2a). Although all plasma and liver biopsy samples tested negative for HCV RNA, three PBMC samples (6%; patients 4, 9 and 20) tested positive (Figure 2b,d). In all three cases, no band was visible after the first round of the RT-PCR reaction (data not shown), which suggests that the initial amount of HCV RNA was very low in these three samples.
......To investigate if HCV RNA persists in the PBMCs of these patients, we took new blood samples 5, 5 and 4 years after the RNA-positive samples. In all three cases, plasma and PBMCs from these blood samples tested negative for HCV RNA (data not shown). This supports the finding that the probability to detect HCV RNA traces in PBMCs of treatment-recovered patients decreases with the time after successful HCV therapy.
........In conclusion, residual HCV RNA can be detected up to 9 years after SVR in a minority of the patients. This low-level HCV RNA may sustain HCV-specific immune responses but does not cause detectable liver disease. Taken together, our data indicate that a treatment-induced SVR corresponds to a cure and that the clinical significance of any residual trace amounts of HCV RNA seems limited."
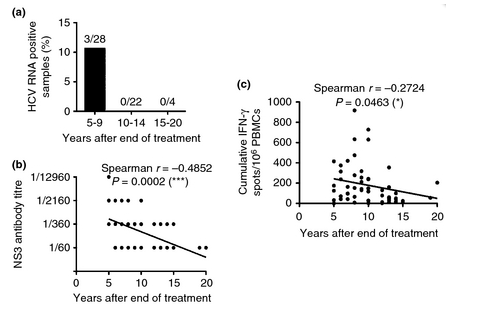
Figure 3. HCV RNA prevalence, B cell activity and T-cell activity decrease with the time after treatment. (a) The analysis of PBMC samples from 54 patients by nested PCR with primers specific for the HCV 5'-untranslated region shows that HCV RNA can only be detected in the first 9 years after the end of treatment. (b) Correlation of HCV NS3 antibody titres and the time after cessation of therapy. The NS3 antibody levels in the plasma were measured by enzyme-linked immunosorbent assay. Antibody titres were determined as the last serum dilution giving an optical density at 405 nm of three times the optical density of the negative control (the same dilution of plasma from a patient who has never been infected with HCV). (c) Correlation of the HCV-specific T-cell activity and the time after cessation of therapy. The HCV-specific T-cell activity was determined by IFN-γ ELISpot. The background (negative control) has been substracted.
----------------------
Long-term follow-up of successful hepatitis C virus therapy: waning immune responses and disappearance of liver disease are consistent with cure
Alimentary Pharmacology & Therapeutics March 2015
M. Hedenstierna*,, O. Weiland*,, A. Brass, D. Bankwitz, P. Behrendt, I. Uhnoo, S. Aleman**, K. Cardell,
A. Fryden, G. Norkrans, A. Eilard, H. Glaumann, T. Pietschmann, M. Sallberg & E. D. Brenndorfer
*Department of Infectious Diseases, Karolinska University Hospital Huddinge, Stockholm, Sweden.
Division of Infectious Diseases, Department of Medicine, Karolinska Institutet, Stockholm, Sweden.
Division of Clinical Microbiology, Department of Laboratory Medicine, Karolinska Institutet, Stockholm, Sweden.
Institute of Experimental Virology, Twincore Centre for Experimental and Clinical Infection Research, Hannover, Germany.
Department of Infectious Diseases, Akademiska University Hospital, Uppsala, Sweden.
**Department of Gastroenterology and Hepatology, Karolinska University Hospital Solna and Huddinge, Stockholm, Sweden.
Department of Infectious Diseases, University Hospital, Linkoping, Sweden.
Department of Infectious Diseases, Sahlgrenska University Hospital, Gothenburg, Sweden.
Clinical Pathology and Cytology, Department of Medicine, Karolinska University Hospital Huddinge, Stockholm, Sweden.
Summary
Background
A sustained viral response (SVR) after interferon-based therapy of chronic hepatitis C virus (HCV) infection is regarded to represent a cure. Previous studies have used different markers to clarify whether an SVR truly represents a cure, but no study has combined a clinical work-up with highly sensitive HCV RNA detection, and the determination of immune responses.
Aim
To determine clinical, histological, virological and immunological markers 5-20 years after SVR.
Methods
In 54 patients, liver biochemistry, histology and elastography were evaluated. Liver biopsies, plasma and peripheral blood mononuclear cells (PBMCs) were tested for minute amounts of HCV RNA. HCV-specific T-cell responses were monitored by ELISpot and pentamer staining, and humoral responses by measuring HCV nonstructural (NS)3-specific antibodies and virus neutralisation.
Results
Liver disease regressed significantly in all patients, and 51 were HCV RNA-negative in all tissues tested. There was an inverse association between liver disease, HCV-specific T-cell responses and HCV antibody levels with time from SVR, supporting that the virus had been cleared. The three patients, who all lacked signs of liver disease, had HCV RNA in PBMCs 5-9 years after SVR. All three had HCV-specific T cells and NS3 antibodies, but no cross-neutralising antibodies.
Conclusions
Our combined data confirm that a SVR corresponds to a long-term clinical cure. The waning immune responses support the disappearance of the antigenic stimulus. Transient HCV RNA traces may be detected in some patients up to 9 years after SVR, but no marker associates this with an increased risk for liver disease.
Introduction
Hepatitis C virus (HCV) infection is one of the leading causes of chronic liver disease and affects 185 million individuals worldwide.[1] About 80% of the patients infected with HCV develop a chronic infection, which can progress to liver fibrosis/cirrhosis, hepatocellular carcinoma and/or liver failure. As a persistent virus, HCV has evolved efficient mechanisms to evade both innate and adaptive immunity.[2, 3] To be able to eradicate the virus, HCV nonstructural (NS)3 together with HCV core-specific immune responses are of particular importance.[4, 5] The standard therapy for chronic hepatitis C based on pegylated IFNα and ribavirin was able to cure 42-84% of the treated patients largely depending on the viral genotype and host interleukin (IL)-28B polymorphisms.[6] Several direct-acting antivirals have been or will soon be approved, resulting in a further improvement of the treatment response and/or safety of HCV therapy.[7]
The treatment goal is to obtain a sustained viral response (SVR) defined as undetectable HCV RNA in serum 24 weeks after the end of treatment, which is regarded to represent a cure from HCV infection. However, several groups have described the presence of HCV RNA traces in the serum, peripheral blood mononuclear cells (PBMCs) or the liver of treatment-recovered patients,[8-11] suggesting that these patients may not have completely eradicated the virus. Furthermore, residual HCV RNA was reported to increase the risk for necroinflammatory activity and fibrosis in the liver,[12] indicating that residual RNA may be involved in liver disease development.
A major concern would be that residual foci of HCV-infected cells with low HCV replication may be present but kept under control by the immune system. If true, this might allow for reactivation of the infection in immunosuppressed patients.
Furthermore, a recent report showed that residual HCV RNA, which appears sporadically in treatment-recovered patients, can be infectious in chimpanzees.[13] However, although HCV reactivation after SVR has been described in case reports,[14-16] this is a truly unusual event.
To better understand the clinical importance of residual HCV RNA after SVR, we screened 54 patients 5-20 years after SVR for HCV RNA traces in plasma, PBMCs and liver, and performed a full clinical and histological work-up. In addition, we evaluated humoral and cellular immune responses to HCV in these patients.
Discussion
In a cohort of 54 patients, we analysed if SVR after chronic hepatitis C therapy represents a clinical cure from HCV. Importantly, all signs of liver disease, HCV-specific T-cell responses and HCV NS3 antibody levels decreased with the duration of SVR irrespective of the transient detection of residual HCV RNA in some patients. Thus, the combined clinical, histological, virological and immunological data support the disappearance of the antigenic stimulus on long-term and confirm that an achieved SVR corresponds to a clinical cure.
Numerous reports have suggested that HCV RNA can persist at low levels in treatment-recovered patients.[8-11] The clinical significance of this has not been clearly determined. In our cohort of treatment-recovered patients, we found residual HCV RNA in 6% of the patients, which is quite similar to the frequency obtained by Morishima et al.,[23] Hoare et al.,[24] and George et al.[25] However, the analysis of successive samples at several time points after SVR in all patients may have resulted in a higher number of HCV RNA-positive patients. Furthermore, repeated sampling of the three HCV RNA-positive patients at more than two time points would have been necessary to investigate if the detection of residual HCV RNA is truly transient or can be detected in consecutive samples.
In two of the three patients with residual HCV RNA, the detected HCV genotype matched the pre-treatment genotype pointing to persistence of autologous HCV RNA. In the third case, however, the pre-treatment genotype was 2b, whereas the detected genotype after SVR was 3a. This patient may have been initially infected with HCV of both genotypes and whereas the 'easier to treat' genotype 2b strain was eradicated, the genotype 3a strain remained. Alternatively, the patient was reinfected with HCV genotype 3a after SVR. We were unable to obtain pre-treatment samples to test these hypotheses.
In all three HCV RNA-positive patients, we detected residual HCV RNA in PBMCs, but not in plasma or liver biopsies. Although some reports describe the detection of negative-strand HCV RNA in PBMCs[26, 27] indicating that low-level HCV replication in PBMCs may be possible, recent studies analysing the association of HCV with PBMCs show that HCV is bound to the surface of PBMCs, in particular B cells.[28, 29] In HCV-infected patients circulating virus can be complexed with immunoglobulins.[30] Thus, immune-complexed HCV may associate with immunoglobulin receptors present on PBMCs. In addition, non-immune-complexed virus may interact with HCV receptors present on the surface of PBMCs. We would favour the idea that the virus associated with the PBMCs does not infect and replicate in PBMC, but is merely bound or associated to the cell surface of some PBMC cell subset(s). Small intrahepatic foci of infected cells, which may explain some cases of HCV reactivation after SVR,[14-16] could be a possible source of the virus. These foci are small in size and number, so that they escaped detection by a liver biopsy.
However, several of our observations suggest that HCV has eventually been completely cleared. First, we did not detect HCV RNA in samples taken more than 9 years after SVR. Second, all three patients who were initially HCV RNA-positive became HCV RNA-negative at later time points similarly as reported by Morishima et al.,[23] Giannini et al.,[31] and Veerapu et al.[11]
The possible low-level HCV replication in the three HCV RNA-positive patients may promote persistence of HCV-specific T-cell responses and the high HCV NS3 antibody levels. This HCV-specific immune response may be responsible for the complete disappearance of HCV RNA on long-term. The low number of cross-neutralising antibodies suggests the presence of a more genotype-specific antibody response in these patients.
In general, the decrease in immunological and liver disease parameters after SVR indicates the clearance of the immune stimulus and suggests a complete cure. We found an inverse correlation between the time after SVR and both HCV NS3-specific antibody titres and T-cell responses. A more homogenous patient cohort regarding the HLA type may have allowed us to find a better correlation of the number of HCV-specific CD8+ T cells using HLA-specific pentamers with the time after SVR. Furthermore, liver disease parameters improved after SVR in all patients. Importantly, the patients with trace amounts of HCV RNA lacked any sign of histological liver disease, suggesting that traces of HCV RNA in the PBMC compartment are not associated with detectable liver disease. This is consistent with results by Radkowski et al.[32] and Carreno et al.,[33] who detected low-level HCV RNA traces in patients with no signs of liver disease.
This study has some limitations. No blood samples were available just after therapy. Therefore no longitudinal comparison could be performed. The group size of three HCV RNA-positive patients is too small to find significant differences between this group and the HCV RNA-negative patient group. Furthermore, patients with different genotypes and HLA types were compared and only few patients had progressed to fibrosis stage F3 or F4 at the time of therapy. While the regression of liver disease after SVR has been reported previously,[34, 35] this study is unique by analysing a complete set of clinical, histological, virological and immunological data from blood and liver samples, which are necessary to judge the clinical relevance of HCV RNA traces after SVR. Furthermore, this is the first study reporting the decrease in HCV-specific T-cell responses after SVR.
In conclusion, residual HCV RNA can be detected up to 9 years after SVR in a minority of the patients. This low-level HCV RNA may sustain HCV-specific immune responses but does not cause detectable liver disease. Taken together, our data indicate that a treatment-induced SVR corresponds to a cure and that the clinical significance of any residual trace amounts of HCV RNA seems limited.
Results
Patient characteristics
A total of 54 patients (29 females and 25 males), who had achieved SVR after HCV treatment, were enrolled. The patients had a median age of 52.1 years at follow-up, the median follow-up time was 9.8 years (range 5-20) post-SVR, and the median duration of infection prior to SVR was 18.4 years. All patients were negative for HCV RNA in serum at follow-up using the routine COBAS AmpliPrep/COBAS TaqMan HCV test. HCV serology and the confirmation test with RIBA were positive for 50/54 patients. The remaining four patients (patients 7, 22, 25 and 28) were reactive in HCV serology but indeterminate (only one HCV specific band) in the confirmation test with RIBA. All four patients had a follow-up time of at least 10 years supporting that antibody titres had decreased over time.
Clinical and histological signs of liver disease wane after SVR
Clinical, biochemical and histological parameters indicating liver disease were compared between samples taken at baseline (before the start of treatment) and at follow-up (Figure 1). Both the ALT (from 136.78 to 24.14 U/L) and AST levels (from 66.66 to 25.14 U/L) in the serum decreased significantly (P < 0.0001, Mann-Whitney U-test) from baseline to follow-up (Figure 1a,b and Table 1). Similarly, we detected a significant decrease (P < 0.0001, Mann-Whitney U-test) of the APRI (from 0.82 to 0.26) and GUCI scores (from 0.89 to 0.27) from baseline to follow-up (Figure 1c,d and Table 1). From 39/54 patients (72%), liver biopsies were available both at baseline and at follow-up and were used to determine the inflammation grade and the fibrosis stage. Both decreased significantly (P < 0.0001, Mann-Whitney U-test) from baseline to follow-up (Figure 1e,f and Table 1). The mean FibroScan elasticity level at follow-up was 4.9 kPa (range 2.4-8.8 kPa) confirming the histological analysis (Table 1). Thus, 5-20 years after SVR, the degree of liver disease had regressed and in the majority normalised according to all measured clinical, biological and histological markers.
Methods
Patients and study design
Patients with SVR after treatment of chronic hepatitis C with interferon or peginterferon alpha-2b ± ribavirin were identified in the participating clinics patient records. These patients had undergone standard pre-treatment clinical evaluation with biochemical and histological testing. 18-65 year old patients, who had achieved SVR at least 5 years prior to inclusion, were offered a follow-up visit for clinical evaluation and histological follow-up with a new liver biopsy and/or liver elasticity testing (FibroScan). At the follow-up visit, blood samples were collected for biochemical, virological and immunological analysis. An additional blood sample was taken after 4-5 years, if patients were HCV RNA-positive at the follow-up visit. Patients were recruited at the Karolinska University Hospital in Stockholm and the University Hospitals in Gothenburg, Linkoping and Uppsala. The patients gave oral and written informed consent to the study, which was approved by the Regional Ethics Committee according to the guidelines of the 1975 Declaration of Helsinki.
Biochemical data and non-invasive fibrosis markers
Biochemical data were collected at the follow-up visit and compared to baseline data defined as data collected within 3 months before the start of the last treatment leading to SVR. Liver fibrosis was assessed by non-invasive serum fibrosis markers such as the GUCI (Gothenburg University cirrhosis index)[17] and APRI (AST to platelet ratio index)[18] scores, which were calculated as follows: GUCI = AST (μkat/L)/ASTULN [upper limit of normal (ULN) AST levels] (μkat/L) x 100/platelet count (109/L) x PK-INR (international normalised ratio) and APRI = AST (μkat/L)/ASTULN (μkat/L) x 100/platelet count (109/L).
Histological methods and liver elasticity measurement
Liver biopsies at baseline (last pre-treatment biopsy) and at follow-up were classified according to grade (A0-3) and stage (F0-4) by using the METAVIR scoring system.[19] The pathologist was blinded to whether the biopsies were taken before or after successful treatment. A liver elasticity measurement was also performed with FibroScan, a non-invasive method to determine the elasticity of the liver, which correlates with the extent of fibrosis/cirrhosis in the liver.[20]
HCV RNA detection
At follow-up, patient serum was routinely screened for HCV RNA by using the COBAS AmpliPrep/COBAS TaqMan HCV test (Roche Diagnostics, Mannheim, Germany) with a sensitivity (95% hit rate) of at least 15 IU/mL for HCV WHO Standard. In addition, HCV serology and Recombinant ImmunoBlot Assay were performed by routine methods. The HCV genotypes have been determined with a line probe assay (Inno-LiPA HCV II; Innogenetics NV, Ghent, Belgium).
A highly sensitive method was used to detect HCV RNA traces in plasma, PBMCs and liver biopsies. Plasma was obtained from heparin-treated blood through centrifugation at 200 g for 5 mins and stored at -80 °C. PBMCs were isolated by using Ficoll-Hypaque density gradient centrifugation and stored in liquid nitrogen. Prior to RNA extraction, patient plasma was concentrated 40-fold through ultracentrifugation at 110 000 g for 17 h by using a Beckman Optima L-80 ultracentrifuge (Beckman Coulter, Brea, CA, USA) and PBMCs (1 x 106 cells per patient) and liver biopsies were homogenised. RNA was extracted by using the TRIZOL method and tRNA (ThermoFisher Scientific, St. Leon-Rot, Germany) as carrier RNA, diluted in 10 μL nuclease free H20 containing RNasin (Invitrogen, Carlsbad, CA, USA) and amplified in a nested polymerase chain reaction (PCR). In a first reaction, RNA was reverse transcribed at 43 °C for 20 min and a 327 base pair fragment of the HCV 5'-untranslated region amplified by PCR by using the outer primer pair Ofor (sense) 5'-CATGGTGCACGGTCTACGAGACC-3' and Orev (anti-sense) 5'-GGCGACACTCCACCATAGATC-3'. In the outer PCR, an initial denaturation at 94 °C for 5 min was followed by 35 cycles of 94 °C for 1 min, 45 °C for 2 min and 72 °C for 3 min, finalised by 5 min at 72 °C. The outer PCR product was further amplified in a second PCR reaction by using the inner primer pair Ifor (sense) 5'-TCGCAAGCACCCTATCAGGCAG-3' and Irev (anti-sense) 5'-GGAACTACTGTCTTCACGCAGA-3' resulting in an inner PCR product of 260 base pairs length. In the inner PCR, an initial denaturation at 94 °C for 1 min was followed by 35 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 1 min, finalised by 3 min at 72 °C. The specificity of the inner PCR product was confirmed by restriction digest with Hae III (ThermoFisher Scientific) resulting in two fragments with 227 and 33 base pairs length. Furthermore, sequencing of the final PCR products was performed (MWG, Ebersberg, Germany).
Detection of NS3 specific antibodies
The specific NS3 antibody titre from human plasma was determined by using a specific solid phase enzyme-linked immunosorbent assay. First, recombinant NS3 (1 μg/mL) in 50 mM sodium carbonate buffer pH 9.6 was passively adsorbed into 96-well plates overnight at 4 °C. Human plasma was serially titrated six-fold with a 1:60 starting dilution and then incubated on the plates for 1 h. Bound antibodies were detected by anti-human IgG-alkaline phosphatase antibody and phosphatase substrate tablets (both from Sigma-Aldrich, St. Louis, MO, USA). Antibody titres were determined as the last serum dilution giving an optical density at 405 nm of three times the optical density at the same dilution of plasma from a patient who has never been infected with HCV.
Detection of neutralising antibodies
Antibodies were isolated from human plasma by using Magne Protein G Beads (Promega, Madison, WI, USA). For inhibition of HCV infection, 200 μL of an Huh7.5 cell suspension (3.5 x 104 cells per mL) were seeded into each well of a 96-well plate 24 h prior to inoculation. Huh7.5 cells were infected with 50 μL SA13/5a/R2a (monocystronic Renilla luciferase virus HCV genotype 5a isolate SA13)[21] in the presence of 36 μg/mL of purified patient antibodies for 72 h. Renilla luciferase activity was measured in a luminometer as described previously.[22] As negative controls, PBS and purified antibodies from control patients and as positive controls, purified antibodies from patients infected with the HCV genotypes 1a, 2a or 3a have been used.
Recombinant proteins and peptides
Recombinant HCV proteins (Core, NS3, NS4, NS5A and NS5B; all corresponding to genotype 1b) were purchased from Mikrogen (Neuried, Germany), Concanavalin A from Sigma-Aldrich. Genotype-specific peptide libraries covering NS3 were used and distributed in four pools with 0.6 μg/mL of each peptide (125 overlapping peptides for genotype 1a, 75 overlapping peptides for genotype 2b, 124 overlapping peptides for genotype 3a).
Human leucocyte antigen (HLA) and IL-28B genotyping
Human genomic DNA was isolated from patient plasma by using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) and for the HLA genotyping further amplified by using the GenomiPhi V2 DNA amplification kit (GE Healthcare, Little Chalfont, UK). HLA genotyping was performed with the Olerup SSP HLA-A and HLA-B typing kits (Olerup SSP, Stockholm, Sweden).
Genotyping for the IL-28B rs12979860 polymorphism has been performed with the TaqMan SNP genotyping assay (Applied Biosystems Inc., Foster City, CA, USA) and the ABI 7500 Fast equipment. The primers and probes used were: Forward primer: 5'GCCTGTCGTGTACTGAACCA3', Reverse primer: 5'GCGCGGAGTGCAA TTCAAC3', Vic probe: 5'TGGTTCGCGCCTTC3' and Fam probe: 5' CTGGTTC ACGCCTTC3'.
Detection of IFN-γ-producing T cells
The human IFN-γ ELISpot was performed according to the manufacturor's instruction (MabTech, Nacka Strand, Sweden) with 0.2 x 106 PBMCs/well. Plates were incubated for 48 h in the presence of recombinant NS3 and NS4 (3 μg/mL each), recombinant core, NS5A and NS5B (3 μg/mL each), four different peptide pools containing overlapping peptides spanning the whole NS3 region (0.6 μg/mL each peptide), medium (negative control) or Concanavalin A (Con A; 2 μg/mL; positive control). Each stimulation was done in duplicates and spot counts were calculated as mean number of IFN-γ spots/106 cells. Cut-offs with ≥50 delta spots (counted spots per 106 cells - spots of the negative control per 106 cells) were used to define significant positive antigen responses. The positive Con A control was >150 SFCs/106 PBMC in all samples included in the analysis proving the viability of the cells. The number of spots was scored using the AID ELISpot reader system Version 7.0 (Autoimmun Diagnostika, Strassberg, Germany).
Detection of NS3-specific CD8+ T cells
The frequency of NS3-specific CD8+ T cells in patient PBMCs was analysed by staining with the pentamers NS3 HLA-A*01:01 ATDALMTGF, NS3 HLA-A*02:01 CINGVCWTV (specific for genotype 1), NS3 HLA-A*02:01 TVGGVMWTV (specific for genotype 3), NS3 HLA-A*24:02 AYSQQTRGL and NS3 HLA-B*35:01 HPNIEEVAL (ProImmune Ltd., Oxford, UK). In brief, PBMCs (1 x 106 cells) were washed and resuspended in PBS/1% FBS (FACS buffer) and incubated with the respective R-PE labelled pentamer for 15 min in the dark at 22 °C. To block Fc binding, cells were washed and incubated with FBS. Cells were washed in FACS buffer and stained with α-human CD19-APC and α-human CD8-FITC for 20 min [antibodies purchased from Becton Dickinson Biosciences (BDB)]. The cells were then fixed in 2% paraformaldehyde in PBS for analysis and 150 000 total events from each sample were acquired on a LSR Fortessa flow cytometer (BDB) using DIVA software. Analysis of flow cytometry data was done in FlowJo version 9.6.2 (Tree Star, Ashland, OR, USA). From a live lymphocyte gate, CD19-positive events were excluded, and the remaining cells were gated for CD8 expression. The frequency of NS3 pentamer positive events within this population was determined.
Statistical evaluation
Nonparametric data were compared using the Mann-Whitney U-test by aid of the InStat 3 software. The correlations after the Spearman's approach were determined by using the GraphPad Prism software. A P-value <0.05 was considered to be statistically significant.
Results
Patient characteristics
A total of 54 patients (29 females and 25 males), who had achieved SVR after HCV treatment, were enrolled. The patients had a median age of 52.1 years at follow-up, the median follow-up time was 9.8 years (range 5-20) post-SVR, and the median duration of infection prior to SVR was 18.4 years. All patients were negative for HCV RNA in serum at follow-up using the routine COBAS AmpliPrep/COBAS TaqMan HCV test. HCV serology and the confirmation test with RIBA were positive for 50/54 patients. The remaining four patients (patients 7, 22, 25 and 28) were reactive in HCV serology but indeterminate (only one HCV specific band) in the confirmation test with RIBA. All four patients had a follow-up time of at least 10 years supporting that antibody titres had decreased over time.
Clinical and histological signs of liver disease wane after SVR
Clinical, biochemical and histological parameters indicating liver disease were compared between samples taken at baseline (before the start of treatment) and at follow-up (Figure 1). Both the ALT (from 136.78 to 24.14 U/L) and AST levels (from 66.66 to 25.14 U/L) in the serum decreased significantly (P < 0.0001, Mann-Whitney U-test) from baseline to follow-up (Figure 1a,b and Table 1). Similarly, we detected a significant decrease (P < 0.0001, Mann-Whitney U-test) of the APRI (from 0.82 to 0.26) and GUCI scores (from 0.89 to 0.27) from baseline to follow-up (Figure 1c,d and Table 1). From 39/54 patients (72%), liver biopsies were available both at baseline and at follow-up and were used to determine the inflammation grade and the fibrosis stage. Both decreased significantly (P < 0.0001, Mann-Whitney U-test) from baseline to follow-up (Figure 1e,f and Table 1). The mean FibroScan elasticity level at follow-up was 4.9 kPa (range 2.4-8.8 kPa) confirming the histological analysis (Table 1). Thus, 5-20 years after SVR, the degree of liver disease had regressed and in the majority normalised according to all measured clinical, biological and histological markers.
Detection of HCV RNA traces in the PBMCs of treatment-recovered patients
Plasma (all 54 patients), PBMCs (all 54 patients) and liver biopsies (40/54 patients) from treatment-recovered patients were tested for HCV RNA in a combined ultracentrifugation/RT-PCR method with a high sensitivity of 2 copies/mL (Figure 2a). Although all plasma and liver biopsy samples tested negative for HCV RNA, three PBMC samples (6%; patients 4, 9 and 20) tested positive (Figure 2b,d). In all three cases, no band was visible after the first round of the RT-PCR reaction (data not shown), which suggests that the initial amount of HCV RNA was very low in these three samples. The specificity of the obtained PCR samples was analysed by Hae III restriction digest (Figure 2c,e) and sequencing. The sequences of the three amplified PCR products differed among the patients, which precludes contamination. Although the pre-treatment HCV genotype matched the post-treatment genotype in patients 9 and 20, the genotype changed in patient 4, in which the pre-treatment genotype was 2b and the post-treatment genotype was 3a. This suggests either a double infection at start of therapy, or a reinfection.
The patients 4, 9 and 20 did not have similar characteristics, such as gender, age, HCV genotype, IL-28B genotype or route of infection (Table 2). However, these three patients had in common that the time between the end of HCV therapy and the follow-up sample was 9 years or less (8, 5 and 9 years) (Figure 3a and Table 2). To investigate if HCV RNA persists in the PBMCs of these patients, we took new blood samples 5, 5 and 4 years after the RNA-positive samples. In all three cases, plasma and PBMCs from these blood samples tested negative for HCV RNA (data not shown). This supports the finding that the probability to detect HCV RNA traces in PBMCs of treatment-recovered patients decreases with the time after successful HCV therapy.
Surprisingly, the three patients with residual HCV RNA in PBMCs only had a very mild liver disease in the follow-up (Table 2). Patient 4 had an ALT level of 64.12 U/L, an AST level of 71.18 U/L, an APRI score of 0.66 and a GUCI score of 0.66, which were slightly above the normal range. However, both the inflammation grade and fibrosis stage were at 0. Patients 9 and 20 were completely healthy according to all liver markers (ALT levels of 18.24 and 7.06 U/L, AST levels of 22.94 and 17.65 U/L, APRI scores of 0.23 and 0.16, and GUCI scores of 0.23 and 0.17, respectively). The biopsy from patient 9 had an inflammation grade of A0 and a fibrosis stage of F0. Unfortunately, no liver biopsy was available from patient 20, but liver elasticity measurement with FibroScan showed a mean value of 4.7 kPa corresponding to a fibrosis stage of F0-1. Overall, this suggests that HCV RNA positivity in the PBMC compartment is not associated with significant liver disease.
HCV NS3 and cross-neutralising antibodies persist long after SVR
To evaluate the humoral immune response in these patients, we determined the NS3 antibody titres and the presence of cross-neutralising antibodies in patient plasma. An NS3 antibody endpoint titre of at least 1/360 was detectable in 29/54 patients (54%) up to 15 years after successful HCV therapy (Table S1).
Importantly, we found an inverse correlation between the NS3 antibody endpoint titre and the time after SVR (P = 0.0002) (Fig. 3B). This suggests that successfully treated patients can maintain HCV antibodies for up to 15 years after SVR but that the levels of HCV antibodies decrease with the time after SVR. Furthermore, this supports the notion that the virus has been eradicated.
The three patients with trace amounts of HCV RNA in PBMCs showed diverse NS3 antibody levels (endpoint titres of 1/2160, 1/2160 and 1/60) (Table 2), indicating that there is no clear association between the presence of trace amounts of HCV RNA and HCV antibody levels. However, very high NS3 antibody titres (≥1/2160) were only seen in eight of the 54 patients (Table S1), and two of these (25%) had trace amounts of HCV RNA. This suggests that a low degree of HCV antigens resulting in antigenic stimulation to maintain HCV antibody levels may exist.
Furthermore, we measured the capacity of antibodies purified from patient plasma to neutralise the infection of hepatoma cells by a nongenotype-matched HCV strain (genotype 5a). Cross-neutralising antibodies with a neutralisation capacity of at least 25% were present in 33/54 patients (61%) (Table S1). Surprisingly, there was no correlation between virus neutralisation and the time after SVR. The three patients with trace amounts of HCV RNA in PBMCs all had low levels of cross-neutralising antibodies (20.5, 9 and 8% cross-neutralisation capacity) (Table 2).
HCV-specific T-cell responses decrease with the duration of SVR
To analyse the HCV-specific T-cell response in these patients, we determined the number of IFN-γ-producing T cells after stimulation with HCV antigens by ELISpot and the number of NS3-specific CD8+ T cells by pentamer staining. 39/54 patients (72%) had at least 50 cumulative spots per 106 cells up to 20 years after successful HCV therapy (Figure 4a and Table S1). We could find an inverse correlation between the number of cumulative spots per 106 cells measured by IFN-γ ELISpot and the time after SVR (P = 0.0463; Figure 3c). Thus, successfully treated patients may maintain HCV-specific T-cell responses for up to 20 years after HCV eradication but the magnitude of the responses decreases with the time after SVR.
The three patients with trace amounts of HCV RNA in PBMCs all had detectable HCV-specific T-cell responses (152.5, 415 and 130 cumulative spots per 106 cells; Figure 4a and Table 2). This suggests that trace amounts of HCV RNA may generate enough HCV antigens to stimulate HCV-specific T cells. However, it should be noted that some patients with undetectable levels of HCV RNA had equally strong or even stronger HCV-specific T-cell responses. This indicates that additional factors are involved in the maintenance of the HCV-specific T-cell responses.
By pentamer staining, we could detect CD8+ T cells specific for known NS3 antigens (at least 0.1% pentamer-positive CD8+ T cells) in 16/41 patients (37%) (Figure 4b and Table S1). We used pentamers specific for HLA-A1, -A2, -A24 and -B35. However, because of alterations in the epitope sequences between the different HCV genotypes, it is probable that not all NS3-specific CD8+ T cells could be detected. Out of the three patients with trace amounts of HCV RNA in PBMCs, patient 9 had 0.42% CD8+ T cells positive for the genotype 1 NS3 HLA-A2 pentamer (Figure 4b and Table 2).
|
|
| |
| |
|
|
|