| |
Differences in disease progression in a cohort of long-term non-progressors after more than 16 years of HIV-1 infection
|
| |
| |
AIDS: Volume 18(8) 21 May 2004 pp 1109-1116
Rodés, Berta; Toro, Carlos; Paxinos, Ellena; Poveda, Eva; Martinez-Padial, Manuelb; Benito, José Miguel; Jimenez, Victoria; Wrin, Terrib; Bassani, Sylvina; Soriano, Vincent
From the Department of Infectious Diseases, Hospital Carlos III, Madrid, Spain, aVirologic Inc, South San Francisco, California, USA, and the bMicrobiology Department, Hospital Carlos III, Madrid, Spain.
Abstract
Background: It is unclear whether resistance to immunologic damage in long-term non-progressors (LTNP) will last indefinitely or whether it merely represents the extreme of a Gaussian distribution, and therefore progression will occur eventually.
Patients and methods: A cohort of 19 LTNP was established in 1997. Plasma viraemia and CD4 cell counts were measured two to three times each year until 2003. Analyses of nef and vpr viral genes, CCR5 genotypes, co-receptor tropism, viral replication capacity, and immunological parameters were performed.
Results: Twelve subjects (non-progressors, NP) showed stable CD4 cell counts over the 6-year follow-up, while seven (slow progressors, SP) showed a trend towards progressive CD4 cell depletion; however, only three SP experienced significant CD4 cell count declines. All SP had detectable plasma HIV-RNA (median 1118 copies/ml). In contrast, five of 12 NP had always undetectable viraemia. Only one patient showed a deletion in nef. The vpr R77Q change was recognized in seven patients. All patients were infected with R5 viruses. The virus replicative capacity was reduced in all tested individuals (range 5-93%). None of the patients was homozygous for the delta-32 CCR5 genotype, which was found in heterozygosis in three. CD8 T-cell activation was low in all but three individuals, all of whom had detectable viraemia and showed progressive CD4 cell depletion. Cytotoxic T lymphocyte responses were similar to those found in a control group of HIV progressors.
Conclusions: A substantial proportion of LTNP show low-level virus replication and progressive loss of CD4 T cells over time. Progressive immunologic damage seems to be directly associated with some degree of virus replication and T-cell activation.
Introduction
The course of HIV-1 infection varies greatly among infected patients. In a small proportion of individuals (1-5%), HIV-1 seems to be less pathogenic and there is no apparent progression. By definition, these long-term non-progressors (LTNP) remain asymptomatic, with CD4 cell counts > 500 ¥ 106 cells/l and low or undetectable viral load, in the absence of any antiretroviral therapy. The cause of the lack of progression in these persons is unclear, but it seems to result from the interaction between multiple factors linked to either the virus or the host. The recognition of what factors are the main determinants for protection against HIV-1 disease progression is of great interest, since it may allow new treatment strategies to be designed.
Strain attenuation has been highlighted as one of the main viral factors accounting for the lack of HIV-1 disease progression. A defective nef gene may impair virus replication, as demonstrated in a cluster of transfusion-infected individuals from the Sydney Blood Bank Cohort. On the other hand, a R77Q change in the vpr gene reduces dramatically cellular apoptosis, which could influence disease progression. Critical changes at other HIV-1 genes such as vif, vpu or env may also account for the lack of progression in some HIV-infected persons. Finally, T-tropic (X4) strains have been linked to increased cytopathogenicity resulting in enhanced T-cell depletion which is lower for M-tropic (R5) viruses.
Among host related factors, some mutations in cellular HIV co-receptor genes have been associated with slower disease progression, as result of an impairment in virus attachment and infectivity. Disease progression is also influenced by the host immune response. HIV-specific cytotoxic T lymphocytes (CTL) play an important role in the control of virus replication, but their relative contribution in LTNP is still uncertain. The role of a limited immune activation or certain MHC haplotypes in HIV-1 disease progression remains also unclear. Whether all of these factors may occasionally result in a truly protective status against HIV-1 disease progression, or whether they merely delay disease progression is not known. Evidence in favor of further damage has been reported recently in small series of LTNP followed for long periods of time, but this does not exclude the possibility that infection could be non-pathogenic in some individuals.
Here we describe the main features of a cohort of LTNP established in 1997 and present longitudinal virologic and inmunologic follow-up for a 6-year period.
Study population
A cohort of 19 patients with evidence of non- progressive HIV-1 infection was established in 1997 at our institution, a reference HIV/AIDS center located in Madrid. All patients had serologically proven HIV-1 infection for at least 10 years, repeated CD4 cell counts > 500 ¥ 106 cells/l, and no prior history of HIV-related symptoms, in the absence of any antiretroviral therapy. Most of these individuals had been exposed to HIV-1 before 1985 or 1987, when they first tested positive for HIV-1 antibodies. One subject was known to be HIV-1 seropositive since 1979, after testing retrospectively sera stored from that time. All of these individuals have been prospectively followed every 4-6 months since January 1997. The last clinical and laboratory assessment was performed in December 2002. The bDNA assay (Quantiplex v3.0, Bayer, Barcelona, Spain), which has a lower detection limit of 50 HIV RNA copies/ml, was used for measuring plasma viraemia, while the CD4 cell count was determined by flow cytometry using specific mAb (Coulter, Barcelona, Spain).
Genetic analyses
Proviral DNA extracted from peripheral blood mononuclear cells (PBMC) was used to amplify nef and vpr genes by nested PCR. Primers and conditions were as described previously. DNA sequencing analysis of the amplified product was performed using an automated sequencer (ABI 3100, Applied Biosystems, Palo Alto, California, USA). DNA and encoded protein sequences were aligned using ClustalX software. Sequences generated in this study were submitted to GenBank and given the accession numbers AY444304-AY444318 for nef and AY444319-AY444336 for vpr.
The presence of the D32-CCR5 allele was determined by PCR in genomic DNA extracted from PBMC. Assay conditions have been described elsewhere. PCR products were analysed in a 2% agarose gel.
Virus replicative capacity and viral tropism
The replicative capacity of HIV-1 was measured by a single-cycle phenotypic assay (Phenosense, Virologic Inc., South San Francisco, California, USA). Results were expressed as percentage of luciferase activity generated by each patient recombinant vector relative to that generated by a wild-type reference virus. Values ranged from 5% (low replicative fitness) to 120% (high replicative fitness).
Viral co-receptor tropism was determined using a modification of the Phenosense assay. Recombinant viruses were tested for their ability to infect CD4/CXCR5 and/or CD4/CXCR4 cells. Tropism was verified by assessing the ability of a CCR5- and/or CXCR4-antagonist to block HIV-1 infection.
Results
Demographics and clinical follow-up
Most individuals in this cohort were first identified as HIV-1 seropositive in 1986 and 1987, and since then have been on regular follow-up at our institution, with periodic controls two to three times per year. They represent a unique subpopulation (approximately 1%), among a total of around 2100 different HIV-infected individuals on regular follow-up at our institution. Eleven (57.8%) of the 19 patients were male. All but one (97.7%) were former injecting drug users. Their mean age was 40.4 years and the mean length of proven HIV-1 infection was 14.7 years in 1997, at the time of cohort establishment. Their mean alcohol intake was 106 g/day (range 0-300 g/day). All were seropositive for hepatitis C virus (HCV) antibodies and 17 patients had markers of prior hepatitis B virus (HBV) exposure. One subject had persistent HBV surface antigen and therefore had been diagnosed as having chronic hepatitis B. Four patients cleared HCV while the rest maintained positive HCV RNA. Interestingly, those four HCV PCR-negative patients belonged to the non-progressor (NP) group.
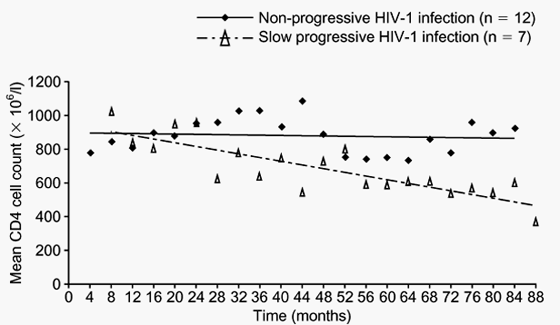
Figs 1 and 2 show the evolution of CD4 T cell counts and plasma viraemia in all 19 LTNP of this cohort during the period between January 1997 and December 2002. Twelve subjects showed stable CD4 cell counts (Table 1), and were classified as NP, while seven subjects showed a reduction in the number of these cells and were classified as slow progressors (SP). The median loss of CD4 T cells in SP was 64 x 106 cells/l per year (Table 2). In three of these SP, the slope of the CD4 decay was statistically significant. All seven SP had detectable plasma viraemia with an average of 1118 HIV RNA copies/ml. In contrast, five out of 12 NP had undetectable viral load at all time points (Table 1). Overall, the mean plasma HIV RNA in NP was 85 copies/ml. The difference in viral load between NP and SP was statistically significant (P = 0.017).
There was no association between the degree of progression (NP or SP) and any of the variables associated with disease progression (e.g., age, sex and high alcohol intake) or with chronic hepatitis B and/ or C.
Fig. 1. Evolution of mean CD4 cell counts in a cohort of 19 LTNP over 6 years of follow-up. Patients are stratified according to the course of their HIV-1 infection.
|
|
| |
| |
 |
|
| |
| |
Fig. 2. Evolution of plasma viraemia in a cohort of 19 LTNP over 6 years of follow-up. Three viral load determinations per year from each patient are represented. Patients are stratified into two categories, according to their HIV-1 disease progression.
Genetic analyses
All 19 LTNP carried HIV-1 subtype B strains. Analysis of nef showed that all but one had a functional nef gene, with no alterations or defects in their functional domains (Table 3). In this patient (number 3) molecular analysis revealed a deletion of 199 bp. This deletion removed the highly conserved acidic domain, (PXX)4 and PKC motifs, and placed downstream sequences out of frame, which resulted in a premature stop codon. This patient has maintained stable CD4 T-cell counts for more than 16 years. No duplications of the nuclear factor (NF)-kB binding elements that could compensate this deletion were found. All of the patients with a complete nef gene showed some amino acid features (T15, N51, H102, L170 and E182) which previously have been associated with non-progressive HIV-1 infection. In fact, their 'Nef progression score' was within the range assigned by others [16] to NP. However, no differences were observed when comparing NP and SP in this study.
Genetic analysis of vpr showed that all 19 LTNP carried a functional gene. The Vpr peptide was 96 amino acids long in all but one individual, who had an insertion of three amino acids at the C-terminal end. The R77Q change was found in seven (36.8%) patients. There were no significant differences when comparing NP and SP: 4/11 (33%) and 3/7 (42.5%), respectively. However, patients with detectable plasma viraemia and/or higher CD38 expression tended to show R77Q more frequently than patients with undetectable viral load and minimal immune activation.
No patient was homozygous for the D32 CCR5 genotype, which was found in heterozygosity in only three subjects (nos. 14, 15 and 17). All patients with D32 CCR5 were SP (Table 3).
Virus replicative capacity and co-receptor usage
The replicative capacity (RC) could be measured in viruses from only seven individuals (three NP and four SP) from whom samples were available. Overall, RC values were lower in LTNP with respect to wild-type viruses taken as controls. However, there was a wide variability in the RC among isolates from LTNP, ranging from 5% to 93%. The median RC values were 40% in NP and 74.5% in SP.
Viral co-receptor tropism was examined in the seven patients tested for RC. All individuals were infected with R5 viruses. However, longitudinal samples collected from one patient (number 15) belonging to the SP group showed a shift from exclusively R5 viruses at baseline to a mixed X4/R5 virus population in samples collected on follow-up. This switch preceded a CD4 T-cell decline of 80 x 106 cells/l per year. Interestingly, virus tropism characterization of the two most recent blood specimens collected from this patient show only R5 viruses.
Activation of T-cell subsets
Levels of CD8 T-cell activation were low in all LTNP but three, all of whom had detectable viraemia and showed a progressive CD4 T-cell decline. There was a trend towards an association between T-cell activation and disease progression. Patients belonging to the SP group showed slightly higher activation of CD8 T cells in respect to NP, although the difference did not reach statistical significance (P = 0.08), probably due to the small size of the study population (Table 3). Activation of CD4 T cells was also lower in LTNP than in controls, but there were no significant differences between NP and SP.
HIV specific CTL responses
The mean number of peptide pools to which patients responded was similar (4 ± 1) in LTNP and a group of controls (HIV progressors). For each pool of peptides, a positive CTL response was detected in LTNP and controls, respectively: 93% and 86% to Gag; 92% and 68% to Pol; 93% and 81% to Env; 87% and 100% to Nef; 40% and 47% to regulatory peptides.
There were no differences in the level of CTL responses against each pool, nor in the level of total responses between LTNP and controls: Gag, 0.52% versus 0.82%; Pol, 0.37% versus 0.43%; Env, 0.32% versus 0.31%; Nef, 0.46% versus 0.53%; Reg, 0.07% versus 0.14%; total, 1.84% versus 2.23%. In both groups, the peptide pool that contributed most to the total CTL response was Gag (28.7% in LTNP versus 32.8% in controls), followed by Nef (23.9% versus 28.7%), Env (22.2% versus 17%), Pol (21.3% versus 13%) and lastly regulatory peptides (3.7% versus 8.2%), respectively. Finally, there were no significant differences in the CTL response when comparing NP and SP.
Discussion
We have described the main features and subsequent outcome of a cohort of 19 LTNP established in 1997. Most of these individuals were first recognized as HIV-1 seropositive in 1985 and 1987, although we have evidence of infection in one of them since 1979. Thus, at the time of cohort establishment all of these subjects had been infected with HIV-1 for more than 10 years. An important finding in our study was the recognition of a different outcome in the study population during the subsequent 6 years of follow-up. Twelve subjects (NP) have remained asymptomatic with stable CD4 T-cell counts while the other seven (SP) have experienced a significant decline of these cells. A similar delayed diminution of CD4 T cells has been reported in other LTNP. We could not find any association of this phenomenon with some of the potential cofactors such as age, sex, alcohol intake or other infectious agents, such as hepatitis viruses.
The majority of individuals assigned to the group of NP had undetectable viral load at all time points. On the contrary, all subjects showing slow progressive disease had detectable levels of viraemia. Thus, the ability of the virus to replicate with more or less efficiency was clearly one critical determinant of the distinct outcome seen in this cohort of LTNP, as has been suggested by others. Although the virus replicative capacity was reduced in almost all of the patients, slightly lower values were observed in NP than in SP. These findings are in agreement with a causal relationship between viral replication and CD4 T-cell depletion. Whether it results from direct cell killing by the virus and/or from indirect mechanisms such as immune activation is open to debate. Most subjects in this cohort of LTNP showed low T-cell activation, which tended to be lower in NP than in SP. Thus, it might be argued that low immune activation could have contributed to preserve these patients from CD4 T-cell depletion.
Specific HIV CTL responses are important in restricting viral replication and probably also in delaying disease progression. However, in our study we did not find differences in the level of CTL responses when comparing LTNP with a control group of drug-naive HIV progressors. Thus, our findings do not support a critical role of CTL responses in halting HIV progression. If relevant, qualitative parameters within the CTL response could be the main determinants of HIV control, as suggested by others.
Non-syncytium inducing strains seem to be less deleterious to the host immune system and along with a low replicative capacity, may determine slower progression of HIV disease. In our cohort, six out of seven LTNP maintained R5 viruses during the entire follow-up. The single patient who showed a shift from R5 to mixed R5/X4 viruses at some point, suffered a coincident rapid loss of CD4 T lymphocytes. The emergence of viral variants capable of using an expanded range of coreceptors most probably explained this adverse outcome in this individual.
The D32-CCR5 allele seems to provide a continuous protective effect during the course of HIV-1 infection, reducing the risk of disease progression by 31% when present in heterozygosity. None of our patients presented this allele in homozygosis and only three carried the D32 deletion in heterozygosis. Since all of them belonged to the SP group, our findings do not support a critical role of CCR5 as cause of LTNP. The protection by the CCR5 delta phenotype is lost when the infecting strain is able to induce syncytia. Interestingly, one of our patients with the D32 deletion was the subject who showed a virus with mixed CCR5/CXCR4 tropism. At the time of its appearance, the patient showed a significant CD4 T-cell depletion, perhaps indirectly reflecting a protective effect of D32-CCR5.
We could not find any other virological feature in these LTNP which could distinguish those showing slow progression and those without any progression at all. Large deletions in nef are rarely present in HIV isolates from infected persons, although they seem to be more common in LTNP. Only one of our patients presented a major deletion that rendered a non-functional nef gene. Moreover, specific amino acid changes in Nef, which have been associated with LTNP, did not differ among NP and SP. Likewise, we did not find differences between vpr genes from NP and SP. The change R77Q was present in 33% and 42% of NP and SP, respectively. These percentages are clearly below those reported originally among LTNP, which were in the range of 80% [3]. Interestingly, most LTNP with detectable viraemia in our cohort presented the R77Q change, and it could be hypothesized that the presence of this mutation might have ameliorated T-cell destruction in those individuals despite them harbouring detectable viral load.
In summary, LTNP seem to be a heterogeneous small subset of HIV-1 infected individuals. No unique viral or host factors explain the absence of disease progression. Even very low levels of HIV replication seem to result in progressive CD4 T-cell depletion, although in these patients it may take decades to become manifest. Whether this applies to all LTNP or whether a subset of them will maintain their CD4 T-cell numbers indefinitely is unclear.
|
|
| |
| |
|
|
|