 |
 |
 |
| |
No Novel Adefovir Mutations
|
| |
| |
Reported by Jules Levin
AASLD, Oct 27-31, 2006, Boston, MA
"Meta-analysis Across Adefovir Clinical Trials Demonstrates the Absence of Novel Adefovir-Associated Mutations and Confirms the Role of the rtA181V and rtN236T Mutations in HBV Polymerase in Association with Virologic Failure"
K Borroto-Esoda, MD Miller, and S Arterburn
Gilead Sciences Inc, Foster City, California, USA
Introduction
HBV resistance mutations have been identified for all approved oral antiviral agents used in the treatment of chronic hepatitis B (lamivudine, adefovir dipivoxil, and entecavir)
The rtA181V and rtN236T mutations have been associated with long-term adefovir dipivoxil (ADV) treatment
Recent reports have proposed that other mutations in HBV polymerase may be associated with ADV resistance
AUTHOR CONCLUSIONS
No ADV resistance mutations (rtA181V and/or rtN236T) were observed in the first 48 weeks of therapy.
No changes in HBV POL at positions other than rtA181 and rtN236 were
signifi cantly associated with failure on ADV therapy when adjusting for multiple comparisons (p < 0.0005):
- Mutations at POL positions rtN236T and rtA181V were significant by themselves
- rtA181T mutation was not significant by itself
Patients who had HBV DNA rebound were more likely to develop adefovir-associated mutations than patients who did not rebound (36% vs. 5%).
Background
Virologic outcomes across ADV clinical studies have been defined based on three criteria:
- Genotypic Resistance Mutations (M) = selection of genotypic changes at critical positions rtA181V and/or rtN236T of HBV polymerase (POL) regardless of
HBV DNA level and ALT outcomes
- Virologic Resistance VR = genotypic resistance and virologic rebound
(confirmed ≥ 1 log10 copies/mL increase in HBV DNA from nadir) and/or having never achieved HBV DNA suppression < 3 log10 copies/mL
- Clinical Resistance (M + VR + ALT) = genotypic resistance along with virologic resistance and ALT elevations (ALT > 1 X ULN after normalizing ALT)
Genotypic resistance to ADV emerges slowly (0% at 1 year, 29% at 5 years) with a cumulative probability of virologic resistance of 20% at 5 years1
Objectives
A meta-analysis of baseline (BL) and on treatment samples from patients across four clinical studies was conducted to:
- Confirm the role of the rtA181V and rtN236T mutations in clinical resistance to adefovir dipivoxil
- Screen for and identify other potential ADV-associated resistance mutations
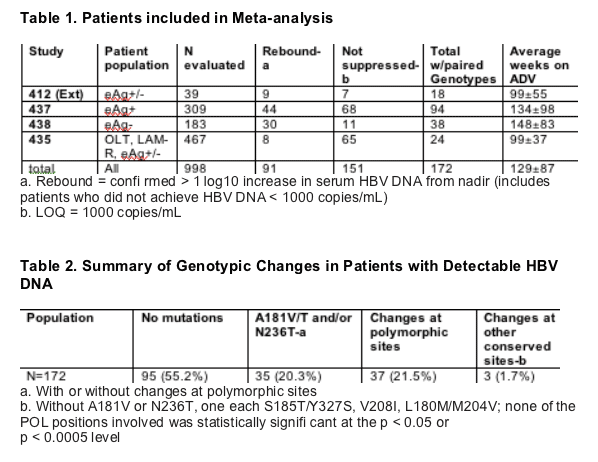
Methods
Patients from the following studies who completed at least 48 weeks of therapy with ADV 10 mg were included in analysis:
- GS-412 (extension), HBeAg positive and negative, naive
- GS-435, HBeAg positive and negative, LAM experienced
- GS-437, HBeAg positive, naive
- GS-438, HBeAg negative, naive
Changes in HBV POL from baseline were determined for the last available genotype for each patient with detectable HBV DNA while still on study medication
Conserved sites were defined as those with only one amino acid observed in the baseline population (n = 853)
Polymorphic positions defined as those with multiple amino acids observed in the baseline populationv
McNemar's Exact test was used to screen paired data (collected at BL and on treatment) for each patient with paired samples across all 4 studies.
The % of patients with a switch from consensus amino acid at BL to non-consensus amino acid while on treatment was compared to the % of patients with a switch from non-consensus amino acid at BL to consensus amino acid while on treatment for each POL position with ≥ 1 patient with a switch (n=97).
p-values used to test for significance were p < 0.05 [no adjustment for multiple comparisons] or p < 0.0005 [Bonferroni adjustment for testing multiple POL positions with changes]
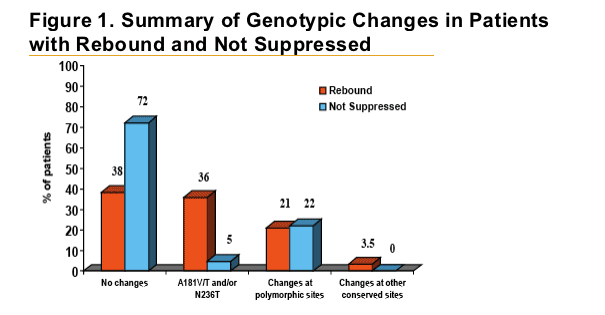
Patients with HBV DNA rebound were almost eight times more
likely to develop ADV-associated mutations at positions rtA181 and/or rtN236 than patients who did not rebound.
Polymorphic changes occurred with similar frequency between both groups and were not statistically associated with a specifi c amino acid change.
|
| |
|
 |
 |
|
|