 |
 |
 |
| |
R1626* (Prodrug of R1479) Demonstrates Potent Antiviral Activity Across HCV Genotypes 1-6 In Vitro
|
| |
| |
Reported by Jules Levin
Presented at the 43rd Annual Meeting of the European Association for the Study of the Liver (EASL), April 23-27, 2008, Milan, Italy
Vincent Leveque, Sophie Le Pogam, Gloria Ao-Ieong, Seth Harris, Hyunsoon Kang, Alan Kosaka, Amritha Seshaadri, Sophie Chiu, Julian Symons, Nick Cammack, Klaus Klumpp and Isabel Nájera, Roche, Palo Alto, California, USA
This research was funded by Roche, Palo Alto, CA, USA
Conclusions
R1479 triphosphate inhibits NS5B enzymes from all six HCV genotypes with similar potency.
R1479 inhibits chimeric HCV replicons from all genotypes tested to date (genotypes 1-4) with similar potency.
Inhibition potency of non-nucleoside inhibitors NNI-1 and NNI-2 were substantially reduced against enzymes and replicons from genotypes 2-6.
The existence of natural NNI-resistant polymorphisms among HCV isolated from treatment-naive patients suggests the potential for faster development of clinically significant resistance to NNI-based therapy as compared with R1479-based therapy.
Introduction
Inhibitors of the NS5B RNA-dependent RNA polymerase interfere with RNA synthesis, a critical step in the replication of hepatitis C virus (HCV).
Two classes of HCV polymerase inhibitors are under development: nucleoside analogs interact with the active site of the enzyme, while non-nucleoside inhibitors (NNIs) bind to different sites on HCV polymerase (Figure 1).
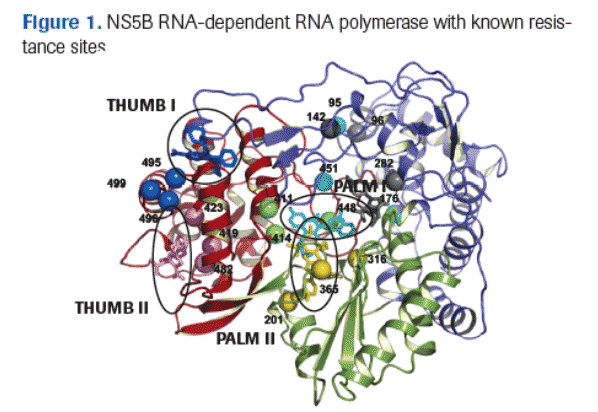
Because nucleoside analogs and NNIs interact with different sites on the enzyme, the resistance profiles of these drugs are expected to be distinct.
A prodrug of R1479, R1626 is being currently evaluated in Phase IIb clinical trials. The nucleoside analog R1479 is a potent and selective inhibitor of HCV replication (Figure 2). R1479 is converted to the HCV polymerase inhibitor R1479 triphosphate by cellular kinases.
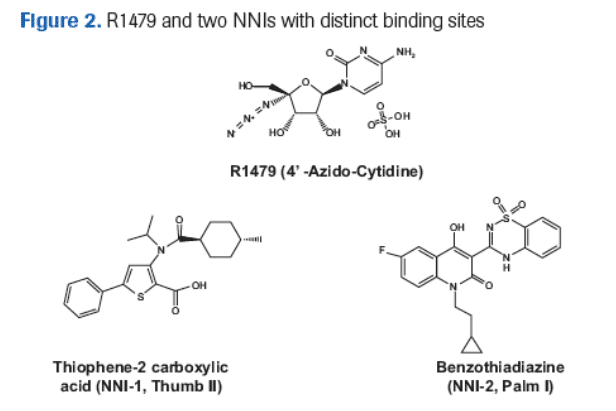
Four weeks of treatment with the combination of R1626 1500 mg twice daily, peginterferon alfa-2a (40KD) (PEGASYS) and ribavirin (COPEGUS) produced a mean maximum reduction in serum HCV RNA of 5.2 log10 in patients with chronic hepatitis C.
The aim of this study was to measure the inhibitory potency of R1479 and select NNIs (Figure 2) against recombinant HCV polymerases and replicons containing NS5B sequences from HCV genotypes 1-6.
Materials and Methods
NS5B clinical isolates
HCV NS5B clinical isolates were obtained from patients infected with HCV genotypes 1-6. After RNA extraction from serum samples, reverse transcriptase-PCR amplification and double- stranded sequencing of the entire NS5B region was performed. NS5B isolates were then cloned in shuttle replicons.1,2 Chimeric replicons from genotypes 5 and 6 did not replicate.
Recombinant NS5B protein from clinical isolates
HCV genotype 1-6 NS5B sequences representing the consensus sequences of each NS5B isolate cloned into the chimeric replicons were selected for recombinant enzyme expression in E. coli. Each NS5B protein was purified to homogeneity.3
Recombinant NS5B J6 protein was purchased from Replizyme Ltd (York, UK).
Public database frequency of natural sequence variability (polymorphism) at the HCV NS5B active site and allosteric NNI binding sites (Palm I and Thumb II)
NS5B sequences were obtained from the Los Alamos database (Table 1).
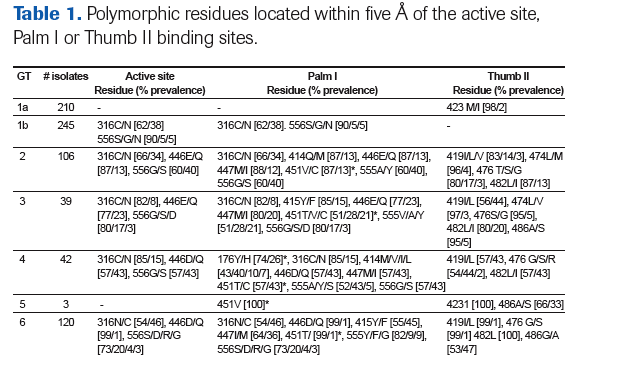
Residues 176 and 451 are located further than five Å of the binding site, but were included in the sequence analysis, because sequence changes at these positions have been previously associated with NNI resistance4
Polymorphism was assessed at each residue within five Å of the active site (Figure 3a), Palm I (Figure 3b) and Thumb II (Figure 3c) binding sites and the frequency was calculated as the percentage of isolates with a particular polymorphism compared to the total number of isolates for each genotype.
A 1% frequency was used as the cut-off for significance.
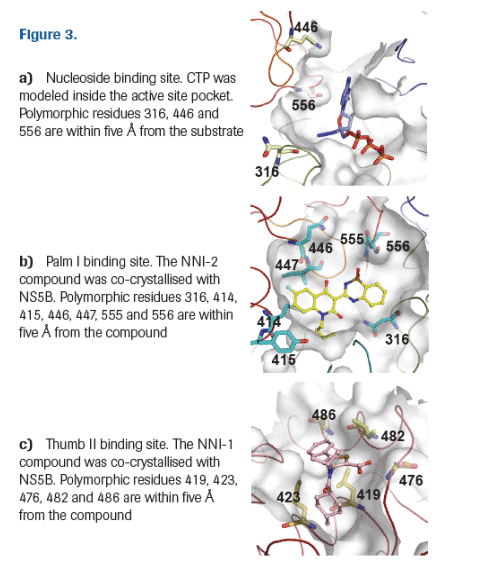
Results
No sequence changes associated with resistance to R1479 were detected in any of the NS5B clinical isolates.
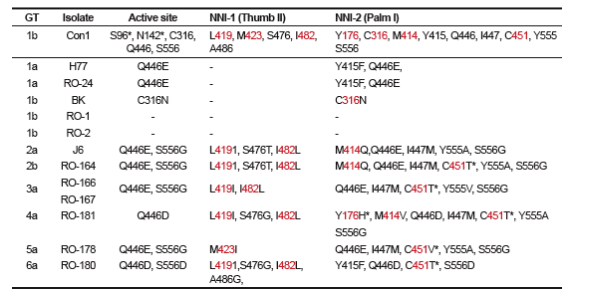
Red text indicates amino acid positions where treatment-induced resistance selection to NNIs has been reported in vitro.4 Although residues 96, 142, 176 and 451 are not located within five ÅA of the binding site, they were included in the table because sequence changes at these positions have been associated with drug resistance in vitro1,4
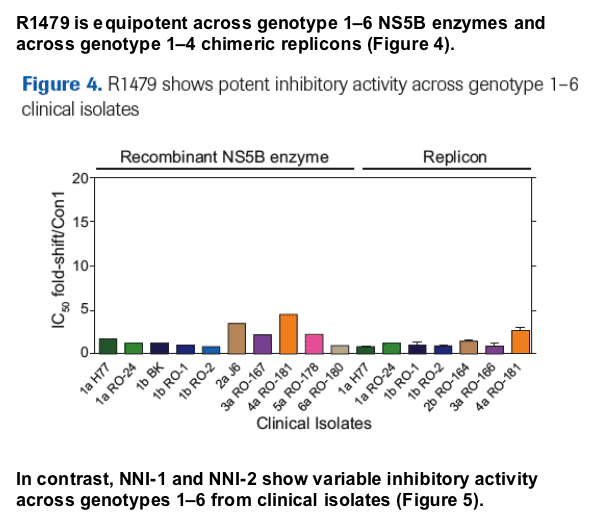
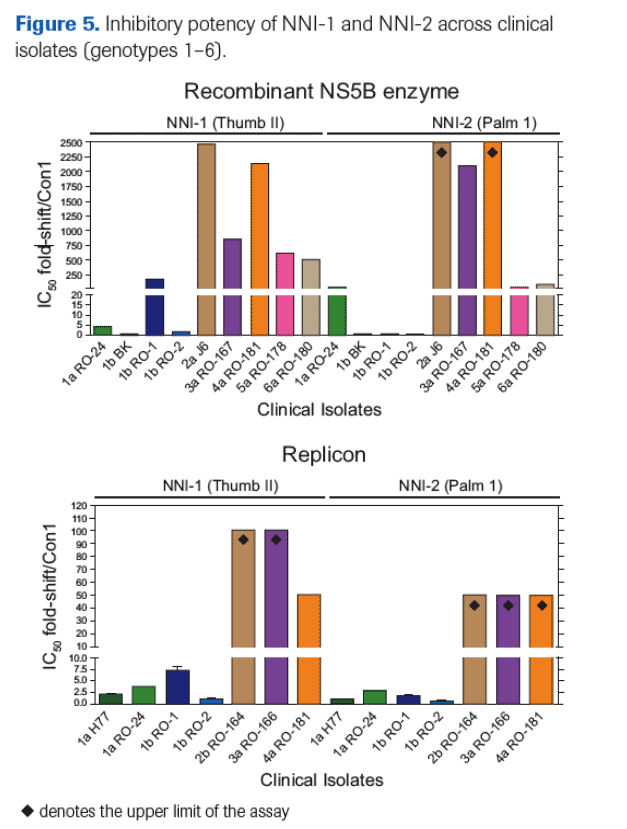
Loss of potency for NNI-1 and NNI-2 correlated with NS5B sequence polymorphisms at positions previously associated with NNI resistance in genotype 1 (Table 2).
High-level natural baseline resistance was detected against NNI-1 and NNI-2 in isolates from multiple genotypes.
References
1. Le Pogam S, et al. In vitro selected Con1 subgenomic replicons resistant to 2'-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 2006;351:349-59.
2. Le Pogam S, et al. Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. JAC 2008 (epub ahead of print).
3. Klumpp K, et al. The novel nucleoside analog R1479 (4'-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. JBC 2006,281:3793-9.
4. Le Pogam S, et al. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J Virol 2006;80:6146-54.
* Prodrug of R1479
|
| |
|
 |
 |
|
|