 |
 |
 |
| |
Incorporation of Optimized Primers into the Trofile Assay Substantially Improves Determination of Viral Tropism in Genetically Diverse HIV Subtypes
|
| |
| |
Reported by Jules Levin
ICAA Sept 11-15 2009 San Francisco
Laura A. Napolitano1, Randy Tressler2, Eoin
Coakley1, Carmeliza Santos1, Jayvant Heera3
and Jeannette Whitcomb1
1Monogram Biosciences, South San Francisco, CA, USA
2Pfizer Inc. New York, NY, USA
3Pfizer Global R&D, New London, CT, USA
BACKGROUND
The HIV epidemic is characterized by broad diversity in HIV subtypes. (Fig. 1)
Recently, global expansion of research studies and improved global access to antiretroviral therapies have increased opportunities to examine a broader range of HIV subtypes.
HIV env, the major determinant of HIV tropism, is characterized by extensive intra- and inter-subtype genetic diversity.
The Trofile assay is a single cycle recombinant virus tropism assay that is validated to detect X4 variants present at levels > 0.3%.1
In the Trofile assay, HIV-1 pseudoviruses are generated using full-length
envelope genes that are derived from RNA PCR amplification of patient virus populations. Coreceptor tropism is determined by measuring the abilities of these pseudovirus populations to efficiently infect cells co-expressing CD4 with either the CXCR4 or CCR5 coreceptor. (Fig. 2)
We interrogated a large database of diverse envelope sequences to optimize primer compatibility for genetically diverse HIV subtypes and increase the likelihood of successful RNA PCR amplification.
The optimized primers were equivalent in overall performance to the original primers, with an expectation of retained performance for subtype B and enhanced performance for non-B subtypes.
METHODS
New primer performance was assessed in cross-sectional prevalence studies designed to examine viral tropism in genetically diverse HIV subtypes in treatment naïve and treatment experienced individuals from India (Pfizer A40001072), South Africa and Uganda (both Pfizer A40001064). (Figs. 3,4)
The established Trofile assay method uses up to 3 mL of plasma and up to 3 attempts at RNA PCR amplification. However, for these study specimens, only a single 1 mL attempt at amplification was made with the original primers. Remaining aliquots were archived for future testing with optimized primers.
Specimens that were non-reportable (NR) using the original primers on the 1st aliquot were re-tested with optimized primers on a 2nd aliquot. If the 2nd aliquot was also NR, optimized primers were used again to test a 3rd aliquot.
The original Trofile assay was used for all tropism testing reported herein. HIV subtype was determined by pol sequencing with the Monogram PSGT assay.
Original and optimized primers were used to determine HIV tropism in > 1000 specimens from treatment experienced patients from S. Africa (subtype C), and in treatment naïve/experienced patients from Uganda (subtypes D, A and A1) and India (subtype C). (Figs. 3,4)
Tropism distribution is reported in Figure 4. Tropism results obtained with the optimized primers were nearly identical to findings from the original primer analysis. (Fig. 4)
Specimens that were non-reportable after 1 attempt with original primers were re-tested up to 2 more times with optimized primers, yielding results in 83% of previously non-reportable samples. This recovery rate of optimized primers was noticeably higher than that ordinarily observed with original primers. (Figs. 5, 6)
Trofile was successful using combined results of original/optimized (vs. original) primers in 98% (vs. 85%) of India subtype C; 93% (vs. 75%) of S. Africa subtype C; and 95% of Uganda samples, performing equally in subtypes D, A and A1 (vs 64% for subtype D; and 84% and 79% for subtypes A and A1, respectively). (Fig. 5)
Analysis of a limited number of aliquot 1 specimens (N=48) suggested equivalent to superior results of the optimized primers in comparison to original primers. (Fig 7)
Thus far, the improved performance of the optimized primers has been confirmed in the following subtypes: Subtype C in South Africa and India, and subtypes A, A1 and D in Uganda.
Assay performance for subtype B is unchanged with implementation of the optimized primers, as measured by reporting characteristics of more than 1000 commercial subtype B specimens (not shown).
CONCLUSIONS
Trofile primers have been optimized to improve compatibility with a broader range of diverse HIV envelope subtypes.
In this analysis of more than 1000 genetically diverse specimens tested with the original Trofile assay, the implementation of optimized Trofile primers considerably improved assay performance with an overall success rate of approximately 95% in HIV subtypes C, A/A1, and D.
Optimized primers have been incorporated into the Trofile assay.
Assay performance for subtype B specimens is unchanged with implementation of the optimized primers.

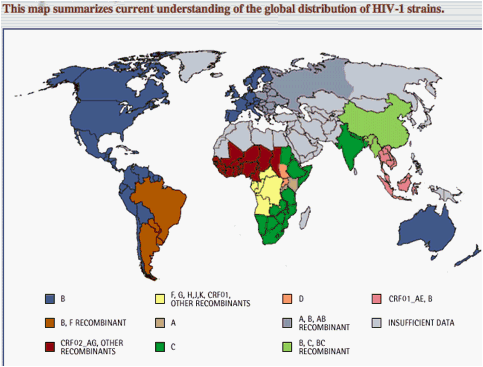
Subtype Prevalence3: C, 50%; A/A1, 12%; B, 10%; G, 6%; AE, 5%; AG 5%;
D, 3%; F,H,J,K < 1% each; all other subypes/CRFs < 0.1% each
Ten different epidemic patterns have been observed, as indicated by the different colors.
In the Americas and Western Europe, subtype B predominates everywhere but eastern South America, where there is a substantial proportion of BF recombinants in addition to subtype B. In eastern Europe, subtypes of A, B, and AB recombinant strains dominate the epidemic. Three different patterns have been observed in Asia: subtype C, a mixture of B, C, and BC recombinants, and a mixture of subtype B and CRF01_AE. The Australian epidemic is subtype B.
Africa shows the greatest diversity. Subtype C dominates the South and East, except for significant foci of subtypes A and D, as shown. West and West Central Africa harbor mainly CRF02_AG, alongside a complex array of other recombinants each present at a low frequency. The most complex epidemic is in Central Africa, where rare subtypes and a wide variety of recombinant forms circulate without any discernable predominant strain.
The map is an overview that does not convey full details of HIV-1 subtypes and recombinants in any given location, and demarcates boundaries more distinctly than they exist in reality. A broad swath cutting across Northern Africa, the Middle East, and Central Asia (gray) is essentially devoid of data on HIV-1 subtypes.
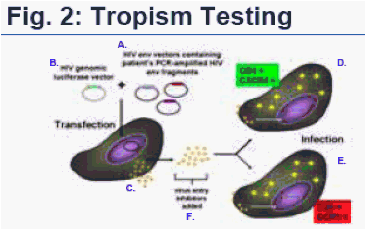
The Trofile Assay: Patient plasma is subjected to RNA PCR amplification of the entire HIV envelope gene (env) to obtain a broad representation of envelope genes from the patient's virus populations. Amplified env products are then inserted into "HIV env expression vectors" (A). The patient's HIV env expression vectors are co-transfected with a full length "HIV genomic luciferase vector" (B) containing a firefly luciferase gene (a fluorescent indicator gene used to quantify viral infectivity) in place of env. Co-transfection of HIV env expression vectors and HIV genomic vectors produces HIV-1 pseudoviruses (C) containing full-length env genes derived from patient virus populations. Coreceptor tropism is determined by measuring the abilities of these pseudovirus populations to efficiently infect target cells co-expressing CD4 with either the CXCR4 (D) or CCR5 (E) coreceptor. Infection is identified by luciferase signal, indicated on the diagram by a large yellow asterisk inside the cells. In the diagram above, both CXCR4+ and CCR5+ cells are infected by pseudoviruses using patient env and, therefore, viral tropism would be classified as 'dual mixed'. To confirm co-receptor usage, additional testing is done: CCR5 and CXCR4 entry inhibitors are added to target cells (F). Viruses susceptible to entry inhibitors will not produce luciferase in target cells.
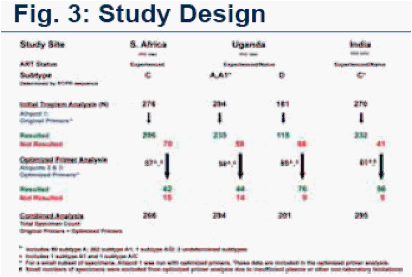
Original primers were used to assess HIV tropism in 1,022 patients from
S. Africa (n=276), Uganda (n=473) and India (n=273). Specimens s that were
non-reportable (NR) with original primers on the 1st aliquot, were re-tested
with optimized primers on aliquot 2 and, if NR, re-tested on aliquot 3.
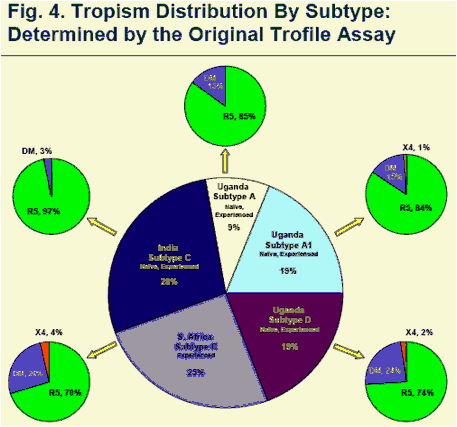
Above data represent all study specimens (N=1022) that produced a tropism result, including specimens tested with optimized primers. Tropism distribution determined with the optimized primers was nearly identical to tropism distribution in the original primer analysis (not shown). Tropism assignment by treatment status in this study was reported recently elsewhere.4

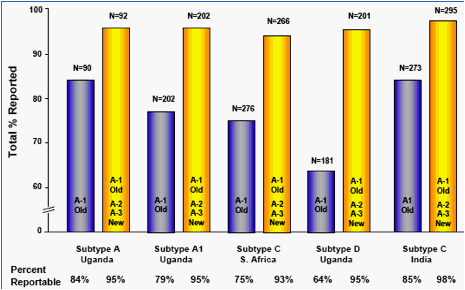
Specimens that were non-reportable using original ("Old") primers on aliquot 1 (A-1), were retested with optimized primers ("New") on aliquot 2 (A-2). If A-2 was non-reportable, optimized primers were used to test aliquot 3 (A-3).
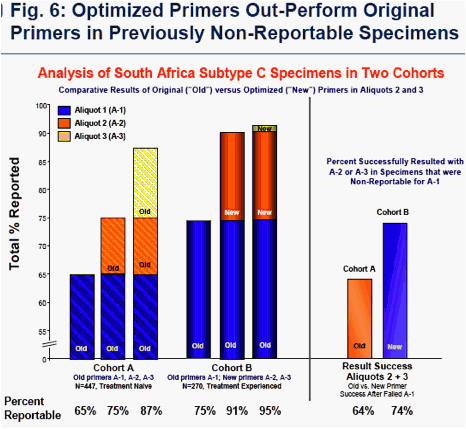
Combined results from all study subtypes (A, D, and C from India and South Africa) established that the overall success rate of optimized primers in aliquots 2 + 3 was 83% in specimens that were non-reportable for aliquot 1

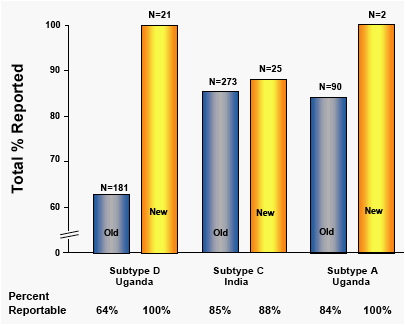
All specimens were aliquot 1 specimens tested with either original ("Old") or optimized ("New") primers
REFERENCES
1. Reeves et al, The Journal of Viral Entry, Vol 3, p. 94, 2009
2. Whitcomb et al. AAC, Vol. 51, p. 566, 2007
3. Taylor et al N Engl J Med, Vol 358, p.1590, 2008
4. Eng et al, 9th International Congress on Drug Therapy in HIV
Infection Glasgow, UK, 2008, Poster P19; Eng et al 5th IAS Cape
Town, S. Africa Poster TUPEA062; Tressler et al, 5th IAS Cape Town,
S. Africa Poster TUPEA063.
|
| |
|
 |
 |
|
|