 |
 |
 |
| |
High -Prevalence of bevirimat resistance mutations
in non-B subtypes and in PI resistant HIV isolates
|
| |
| |
Reported by Jules Levin
18th Intl HIV Drug Resistance Workshop
June 9-12
Ft Myers Florida
J Verheyen1, C Verhofstede2, E Knops1, L Vandekerckhove2, A Fun3, D Brunen3, K Dauwe2, A Weinsing3, H Pfister1, R Kaiser1, M Nijhuis3
1 Institute of Virology, University of Cologne, German
2 Aids Reference Lab, Ghent University, Ghent, Belgium
3 Dept of Medical Microbiology, University Medical Center Utrecht, The Netherlands
BACKGROUND
Bevirimat is the first drug in the class of maturation-inhibitors that hamper the processing of the gag CA/p2 precursor protein. Mutations in the bevirimat target region conferring phenotypic resistance, have been identified after in vitro selection experiments (aa 358/363/364/366). Furthermore, mutations in the QVT-motif (aa 369-371), associated with reduced bevirimat activity, have been observed as baseline polymorphism in several clinical isolates. The objective of this study was to assess the prevalence of bevirimat resistance mutations in isolates from treatment-naïve and PI-experienced patients.
AUTHOR CONCLUSIONS
Mutations conferring resistance to bevirimat accumulate in subtype B isolates with resistance mutations in the viral protease.
HIV non-B isolates display great diversity in the bevirimat taget regio but could be grouped according to gag position 370.
These data emphasize the screening for gag mutations before administration of bevirimat.
The impact of newly detected mutations in the bevirimat target region and of the genetic background in different HIV clades for bevirimat resistance has to be determined in more detail.
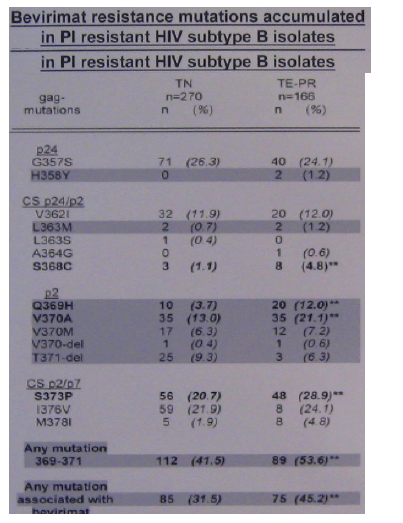
In the 166 patients with PI resistance the accumulation of specific mutations was observed at both cleavage sites (S368C at CS p24/p2 (p<0.05) and S373P at CS p2/p7 (p<0.05)). Furthermore, two mutations in the QVT motif, which were associated with bevirimat resistance, were more frequently identified in the TE-PR patient group (Q369H (p<0.01) and V370A (p<0.05)). Overall, mutations associated with bevirimat resistance (H358Y, L363M, Q369H, V370A/M/del and T371del) were detected in 45.2% of all TE-PR isolates (p<0.01). More detailed analysis revealed a significantly higher frequency of these mutations in TE-PR isolates with three or more PI mutations compared to isolates with two or less PI mutations (TE-PT 1-2 PI mutations 23/67 (34%) vs TE-PR >/=3 PI mutations 52/99 (53%), p<0.05).
METHODS
Of 644 HIV isolates collected at 3 different sites in Europe the bevirimat target region comprising gag aa 357 to 382 was sequenced. Isolates from treatment-naïve (TN) (B: n=270; non-B: n=167) and PI-experienced patients harboring resistance mutations in the viral protease (TE-PR) (B: n=166; non-B: n=41), were selected. Overall, non-B HIV isolates were classified according pol and gag genotypes (A1: 14,, A2: 5; AE: 28, AG: 52, C: 25, D: 7, G: 12, H: 1 and other circulating recombinant forms: 57).
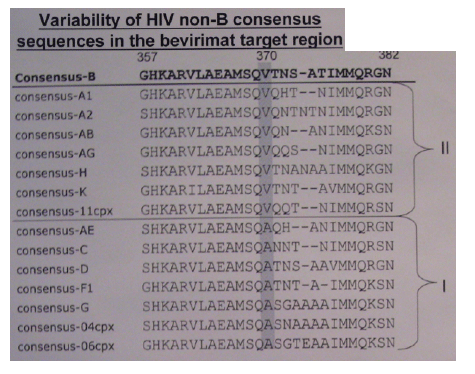
Due to numerous polymorphisms, insertions and deletions in HIV non-B isolates within the bevirimat target region, the consensus sequences of HIV clades were aligned according to the highest homology in the nucleotide sequences. This alignment revealed a specific mutation pattern for each analysed HIV consensus sequence from position 370 to 376. Especially, position 370 was previously shown to influence the susceptibility of HIV isolates to bevirimat, since mutations V370A, V370M and V370- introduced in a HIV subtype B background conferred phenotypic resistance. Therefore, the non-B HIV isolates were grouped according position 370 in their consensus sequences: (group 1 (V370V)= 11cpx, A1, A2, AB, AG, H and K and group II (V370A)= 04cpx, 06cpx, AE, C, D, F, and G).
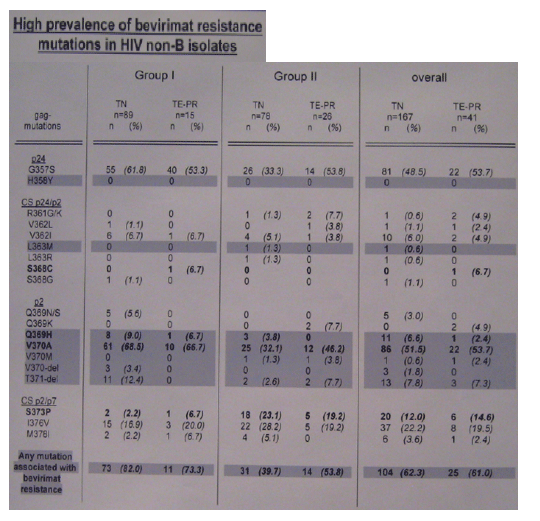
All HIV gag sequences were classified according to the highest homology with one of the compared consensus sequences and separated in two groups (group II = 11cpx, A1, A2, AB, AG, H and K and group I gag mutation V370A was detected leading to at least one bevirimat resistance mutation in about 80% of these isolates. Interestingly, in group II the frequency of mutation V370A increased in TE-PT isolates and more than 50% of these isolates harboured at least one bevirimat resistance mutation. Similar results were found in the group of HIV subtype B isolates and likewise indicated the selection of bevirimat resistance mutations in PI resistant HIV isolates, although statistical significance in no-B isolates was not reached due to the small numbers.
|
| |
|
 |
 |
|
|