 |
 |
 |
| |
HIV Integrase Genotypic and Phenotypic Changes Between Day 1 and Day 11 in Subjects with Raltegravir Resistant HIV Treated with S/GSK1349572: Results of VIKING Study
|
| |
| |
Reported by Jules Levin
18th Intl AIDS Conference, July 18-23 2010, Vienna (home of gelato)
Bonaventura Clotet1, Edwin De Jesus2, Adriano Lazzarin3, Michel Livrozet4, Philippe Morlat5, Cindy L Vavro6, Joseph H Horton6, Jenny Huang6, Akihiko Sato7, Mark R Underwood6
1Hospital Germans Trias I Pujot, Barcelona, Spain, 2Orlando Immunology Center, Orlando, FL, 3Fondazione Centro S. Raffaele del Monte Tabor, Milano, Italy, 4Hopital Edouard Herriot,Lyon, France, 5CHU de Bordeaux-Hopital Saint Andre, Bordeaux, France, 6GlaxoSmithKline, Research Triangle Park, NC, US, 7Shionogi & Co., Ltd., Osaka, Japan
Abstract
Background: Subjects experiencing virologic failure under RAL-containing regimens were enrolled in Viking to assess S/GSK1349572 efficacy, virology, and safety endpoints. We evaluated genotypic and phenotypic changes in HIV IN during 10 days of treatment with S/GSK1349572 plus a failing background regimen.
Methods: IN genotypes and phenotypes were generated for 27 subjects at Day 1 and 18/27 subjects at Day 11 (Monogram BioSciences, Inc. Integrase Geneseq and PhenoSense assays). IN genotype resistance amino acid changes were defined as the gain or loss of RAL-associated resistance substitutions orS/GSK1349572 in vitro passage-associated substitutions, full or mixed, from Day 1 to Day 11. S/GSK1349572 susceptibilities were defined as fold change IC50(FC) versus wild-type.
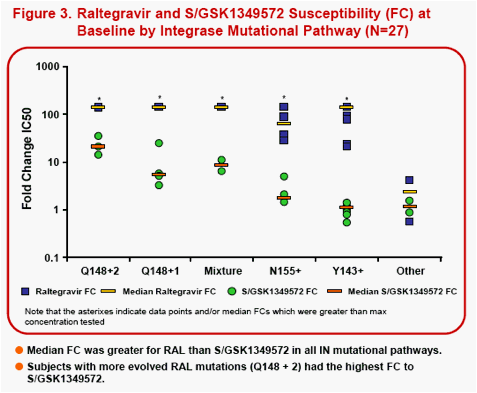
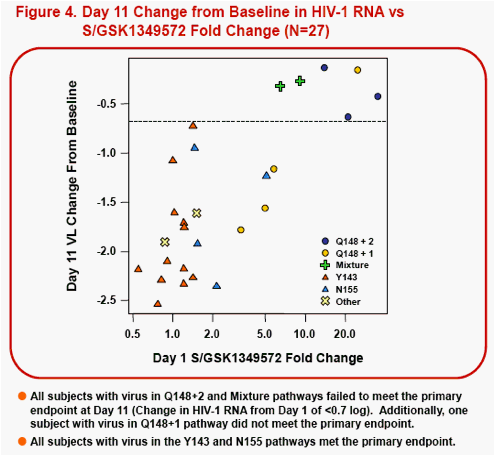
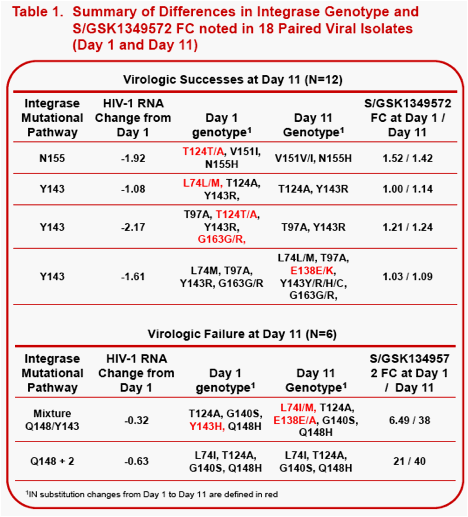
Results: All subjects had RAL signature mutations at screening. On Day 1, 25/27 subjects had virus with RAL-associated signature pathways: Y143 (n=12); Y143+Q148 (n=2); Q148 (n=7); and N155 (n=4). 21/27 subjects at Day 1 had FC <8, 18/27 had FC<4, and 16/27 had FC<2. By Day 11, 5 subjects had genotypic resistance changes. 4/5 had changes in non-signature codons with no susceptibility change to S/GSK1349572: two with Y143-pathway, one with N155-pathway (mutant to wildtype changes); and one with Y143-pathway (wildtype to mutant change). Only one subject with a Day 1 Y143+Q148 (mixed virus) pathway had S/GSK1349572 susceptibility change; FC=6.49 (Day 1) to FC=38 (Day 11) and Day 11 resistance changes including both wildtype to mutant L74I/M,E138E/A and mutant to wildtype Y143H to Y143Y. S/GSK1349572 FC increase is consistent with lack of suppression of viral subpopulations with low S/GSK1349572 susceptibility. An additional subject without resistance changes had S/GSK1349572 Day 1 FC=21 which increased to FC=40 at Day 11.
Conclusions: In RAL-resistant subjects receiving 10 days of S/GSK1349572 50mg with a failing background regimen, few IN genotypic differences were observed, and minimal changes in S/GSK1349572 susceptibility were seen between Day 1 and Day 11, thus demonstrating minimal virus evolution using these analytic methods.
Introduction
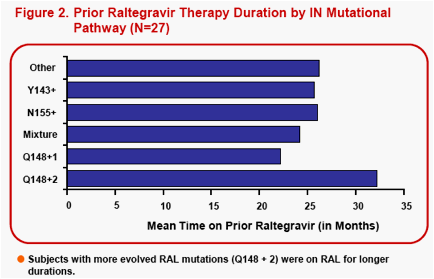
In vitro studies1have shown S/GSK1349572 has limited cross-resistance to RAL and ELV, and good in vitro activity against Q148 single mutant viruses, and against site-directed mutant viruses with Y143 or N155 signature mutations regardless of RAL-associated secondary mutations. However, S/GSK1349572 activity against viruses with Q148 plus additional RAL resistance-associated substitutions has been shown to display a broader range and more reduced activity, particularly as the number of RAL-associated mutations increases2.
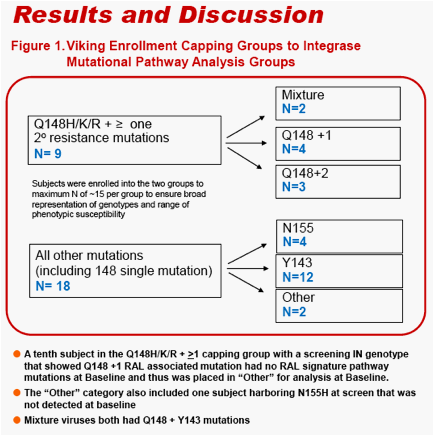
VIKING is a phase IIb pilot study designed to investigate S/GSK1349572 dosed 50mg once daily in ART-experienced subjects with either current or past virologic failure to raltegravir (RAL) harboring virus with RAL signature mutational pathways, Q148, N155 or Y143.
An Integrase screening genotype was used to assign up to 15 subjects into one of two groups.
-- Q148H/K/R plus one or more Q148-associated integrase resistance mutations (at L74 and/or E138 and/or G140).
--N155H and/or Y143 plus any IN mutations, or Q148H/K/R single mutants
Objective: Evaluate genotypic and phenotypic changes in HIV IN during 10 days of treatment with S/GSK1349572 plus a failing background regimen.
METHODS
Results were analyzed according to the following Integrase Mutational Pathway groups.
-- Q148 - Q148H/K/R and no additional Q148-associated resistance mutations
-- Q148+1 - Q148 H/K/R plus one Q148-associated resistance mutation at L74,
-- E138, or G140
-- Q148+2 - Q148H/K/R plus two Q148-associated resistance mutations at L74, E138, and/or G140
-- N155 - N155H w/ or w/o additional IN mutations
-- Y143 - Y143C/H/R w/ or w/o additional IN mutations.
-- Mixture - mutations at Q148H/K/R and Y143C/H/R or N155H
-- Other - mutations present in integrase but not at Y143, Q148, or N155.
Baseline genotypic and phenotypic data were generated by Monogram BioSciences using Integrase Geneseq, Integrase PhenoSense, and PhenoSenseGT (PSGT) assays.
The Abbott RealTime HIV-1 assay was used to generate HIV-1 RNA results.
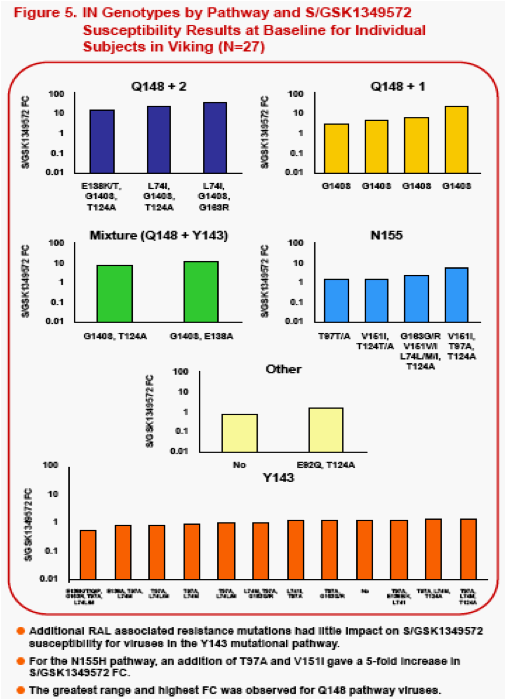
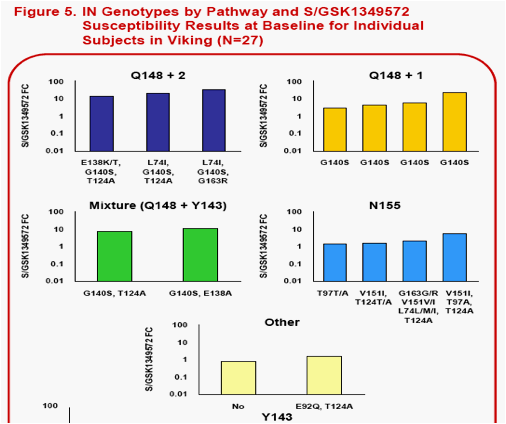
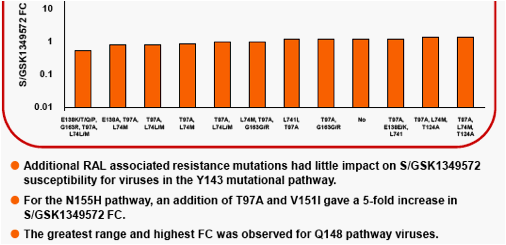
The following Integrase Inhibitor (INI) substitutions were considered in the analysis. RAL signature mutations -Q148H/K/R, Y143C/H/R, and H155H. RAL associated resistance mutations -L74M/I, T97A, E138K/S, G140S, V151I, G163K/R . IN substitutions observed during in vitropassage of S/GSK1349572 -S153Y/F, G193E, E92Q (the latter was also observed in RAL clinical studies) and T124A (polymorphic with no increase to S/GSK1349572 FC).


|
| |
|
 |
 |
|
|