 |
 |
 |
| |
BI 224436, a Non-Catalytic Site Integrase Inhibitor, is a potent inhibitor of the replication of treatment-naïve and raltegravir-resistant clinical isolates of HIV-1
|
| |
| |
Reported by Jules Levin
ICAAC Chicago Sept 17-20 2011
C. Fenwick1, R. Bethell1, M. Cordingley1, P. Edwards1, A-M. Quinson2, P. Robinson2, B. Simoneau1 and C. Yoakim1 1Boehringer Ingelheim Ltd., R & D, Laval, Canada, 2 Boehringer Ingelheim Pharmaceuticals Inc., Ridgefield, Connecticut, USA
Abstract
Background: Non-catalytic site integrase inhibitors (NCINIs) represent a novel class of HIV-1 antiretroviral. They bind to a conserved allosteric pocket on integrase (IN) and have a distinct mechanism of action compared to raltegravir and elvitegravir which bind at the IN catalytic site.
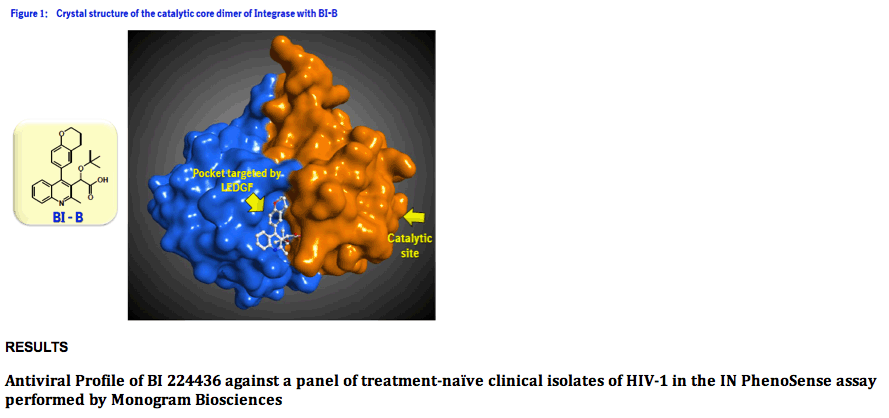
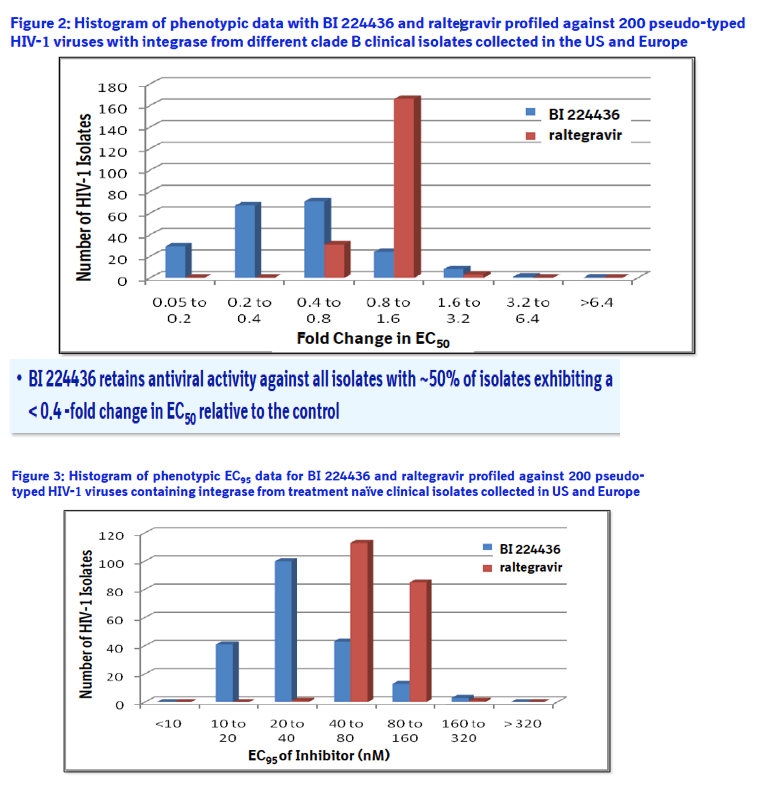
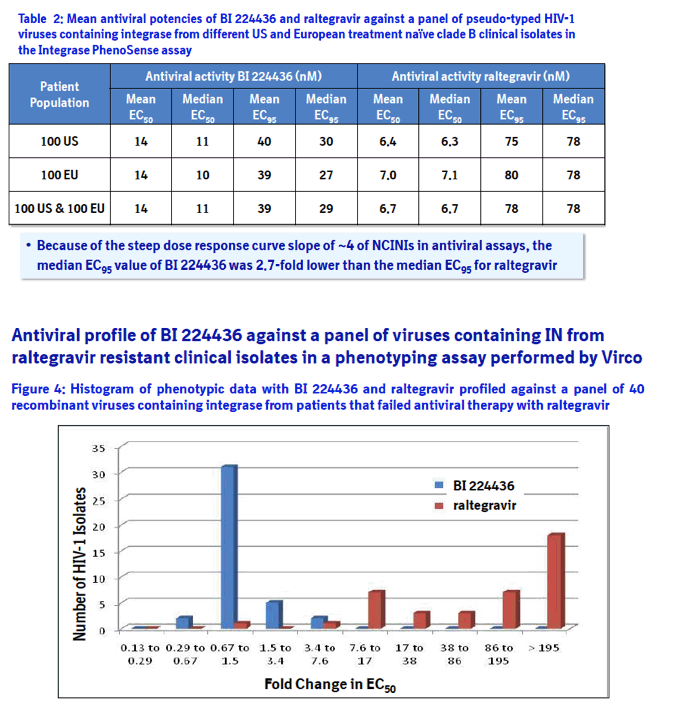
Method: The in vitro antiviral activity of BI 224436 against 200 recombinant viruses encoding the IN from different clade B clinical isolates of HIV-1 was assessed using the PhenoSense assay. Isolates were from 100 US and 100 European patients considered to be treatment naïve based on the absence of NRTI and NNRTI associated resistance mutations. The antiviral activity of BI 224436 was also assessed against a panel of 40 recombinant viruses containing IN from clinical isolates of HIV-1 resistant to raltegravir in an in vitro phenotyping assay performed by Virco.
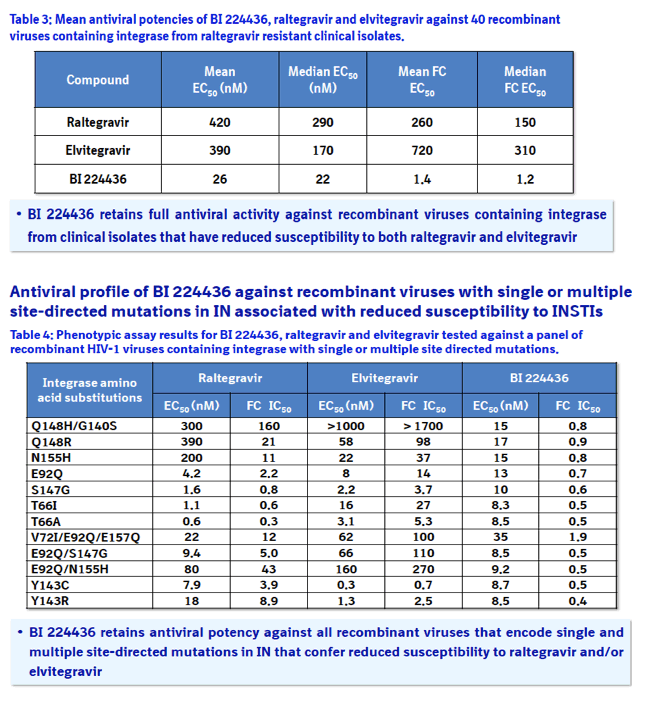
Results: BI 224436 had a mean EC50 value of 14 ± 13 nM for the 200 clinical isolates tested in the PhenoSense assay. Consistent with BI 224436 having a steep dose-response curve slope of ~4.0, the mean EC95 value against these 200 viruses was 39 ± 30 nM. By comparison, raltegravir had an EC50 value of 6.7 ± 1.4 nM and EC95 of 78 ± 18 nM against the same panel. BI 224436 also retained antiviral potency against 40 recombinant viruses with IN from raltegravir resistant clinical isolates. BI 224436 had a mean EC50 value of 26 nM ± 16 nM and mean fold change (FC) in EC50 values of 1.4 ± 0.8 relative to a wild type control virus. Raltegravir and elvitegravir profiled in parallel both displayed a > 260 FC in EC50.
Conclusion: BI 224436 had a potent antiviral profile in IN phenotyping assays against treatment naive clade B isolates of HIV-1 and raltegravir-resistant clinical isolates. Based on its excellent biological and PK profile, and an absence of cross resistance to all other classes of approved antiretroviral agents including IN catalytic site inhibitors such as raltegravir, BI 224436 has been advanced into Phase I clinical trials.
Introduction
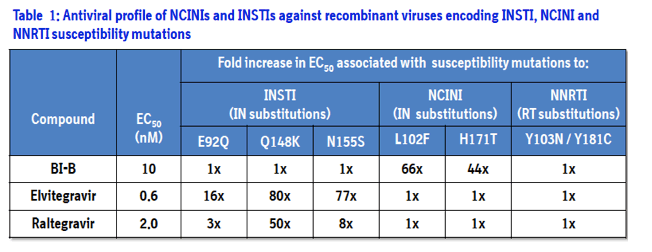
The primary function of HIV integrase is to integrate newly synthesized viral DNA into the host genome, which is an obligate process for subsequent expression of viral genes. Integrase performs two related reactions: the enzyme-catalyzed hydrolysis of a dinucleotide from the 3'-end of each viral LTR DNA and the strand transfer reaction that integrates the viral DNA into the host cell's DNA.
In a high throughput screening campaigns at Boehringer Ingelheim (BI), selective hits were identified using an integrase LTR DNA 3'-processing assay and were sequentially optimized through a combination of medicinal chemistry, parallel synthesis and structure guided drug design.
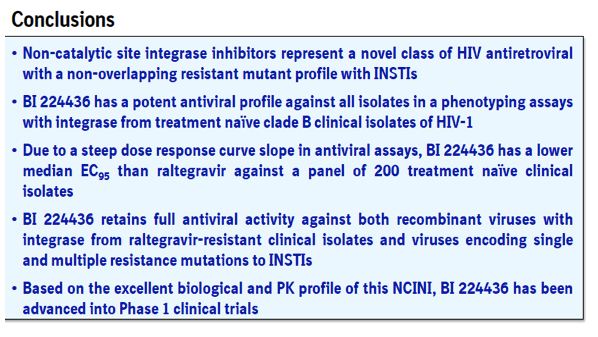
This novel class of integrase inhibitors, referred to as Non-Catalytic site Integrase Inhibitors (NCINI), bind to a conserved allosteric pocket on integrase that is also targeted by LEDGF.1 Compounds from the NCINI class have been shown to maintain antiviral activity against recombinant viruses encoding INSTI or NNRTI susceptibility mutations (Table 1).2 In addition, raltegravir and elvitegravir have been shown to maintain antiviral activity against recombinant viruses encoding substitutions in integrase that result in reduced susceptibility to the NCINI series.
BI 224436 is a potent NCINI antiretroviral that is closely related to BI-B and has been advanced into Phase 1 clinical trials. Further details on BI 224436 are presented in ICAAC posters A1-1725, A1-1726 and F1-1369.
Methods
Phenotyping of HIV-1 integrase from treatment naïve clinical isolates: The integrase PhenoSense assay3 was performed by Monogram Biosciences with 100 US and 100 European treatment naïve clade B clinical isolates selected from a wide geographic distribution throughout the US (patients from: 2 AL, 19 CA, 1 CT, 3 DC, 5 FL, 2 GA, 18 IL, 2 MA, 1MD, 1ME, 1 MI, 2 NC, 3 NJ, 27 NY, 1 TN, 9 TX, 2 VA and 1 WA) and Europe (patients from : 4 Belgium, 13 France, 21 Germany, 1 Ireland, 5 Italy, 2 Netherland, 8 Portugal, 21 Spain, 6 Switzerland and 19 United Kingdom). Fold change in EC50 was determined relative to a NL4.3 control.
Phenotyping of HIV-1 integrase from patients that failed antiviral therapy with raltegravir: Phenotyping was performed by Virco with a panel of 40 viruses selected with resistance mutations in integrase that represent the most prevalent INSTI escape pathways including E92Q, G140S/A, Y143C/R, Q148H/K/R, N155S amino acid substitutions.4 Fold change in EC50 was determined relative to an HXB2 control. In addition, recombinant viruses were generated by introducing single and multiple amino acid substitution in the integrase gene by site directed mutagenesis in the HIV-1 IIIB laboratory strain.
1- Cherepanov, P. et al PNAS 2005 102(48):17308
2- Fenwick, C et. al International Workshop for HIV and Hepatitis Drug Resistance, 2011, Poster #1
3- Petropoulos CJ et al. Antimicrob Agents Chemother. 2000;44(4):920
4- Hertogs K, et al. Antimicrob Agents Chemother 1998;42(2):269.
Other scientists who supported this work include: Murray Bailey, Lee Fader, Anne-Marie Faucher, Stephen Kawai, Marc- Andre Poupart, & Youla Tsantrizos in Chemistry; Pierre Bonneau, Rene Coulombe, Araz Jakalian & Steven LaPlante in Structural Research; Ma'an Amad, Mireille Cartier, Jianmin Duan, Rob Elston, Louie Lamorte, Laibin Luo, Michel Garneau, Steve Mason, Nathalie Rioux & Myriam Witvrouw in Biological Sciences.
|
| |
|
 |
 |
|
|