| |
NS5A Mutations/HCV Drug Resistance
|
| |
| |
Download the PDF Here
Download the PDF Here
EASL: NS5A Mutations/Resistance - (05/11/15)
Letter
Baseline drug resistance mutations are detectable in HCV genes NS3 and NS5A but not NS5B in acute and chronic HIV-coinfected patients
Antiviral Therapy 2015; 20:361-363
doi: 10.3851/IMP2871
Date accepted: 24 June 2014
Date published online: 03 October 2014
Adele L McCormick1,*, Luke Moynihan1, Malcolm J Macartney1, Ana Garcia-Diaz1, Colette Smith2, Margaret A Johnson3, Alison J Rodger2,4, Sanjay Bhagani3,4, Tanzina Haque1, Daniel P Webster1,3
1Department of Virology, Royal Free London NHS Foundation Trust, London, UK
2Department of Infection and Population Health, University College London, London, UK
3Department of HIV Medicine, Royal Free London NHS Foundation Trust, London, UK
4Department of Infectious Diseases, Royal Free London NHS Foundation Trust, London, UK
In 2012, Plaza et al. [1] reported the prevalence of natural polymorphisms in the HCV NS5A gene associated with resistance to daclatasvir (DCV) in 78 HIV-HCV-coinfected patients and 635 HCV-monoinfected derived NS5A sequences deposited in Los Alamos HCV database. They did not observe NS5A resistance-associated variants (RAVs) in HCV-1a and HCV-3 NS5A sequences, whereas, major RAVs (Y93H) were detected in 7% and 13% in NS5A sequences from coinfected patients infected with HCV-1b and HCV-4, respectively, with a similar frequency of NS5A RAVs observed in HCV-monoinfected patients for these HCV genotypes (gts) [1]. Additionally, the L31M NS5A variant was observed in 7% of HCV gt-1b patients, irrespective of coinfection status and occurred in >93% of HCV gt-4 monoinfected and coinfected patients [1]. Interestingly, the presence of naturally occurring drug resistance variants in acutely HCV-infected, treatment-naive HIV patients have been detected by population and deep sequencing in HCV NS3 in a large proportion of subjects [2]. The significance of the threshold at which these RAVs are detectable and whether these will impact on response to therapy with NS3 and NS5A inhibitors in clinical practice is not fully clear. In acute HCV amongst those who are HIV-infected, the role for new HCV direct-acting antivirals (DAAs) has not been established. Treatment with pegylated interferon and ribavirin (PEG-IFN/RBV) early in HCV infection is often successful for most gts [3]. Telaprevir, a first-generation protease inhibitor (PI), has been used in a small study of acute HCV infection in HIV, and results suggest sustained virological response (SVR) may be improved using 'triple therapy' with shortened treatment durations [4]. It is likely that in the near future DAA-based therapy will be standard of care for both acute and chronic HCV infections, with PIs forming part of this armamentarium [5,6] alongside other DAAs, including NS5B polymerase inhibitors [7] and H-CV NS5A inhibitors [8]. Ultimately, an interferon-free future is heralded, where drug regimens will consist of combinational DAAs, targeting different HCV gene products [9]. It is therefore important to establish the frequency of RAVs in all patients infected with HCV.
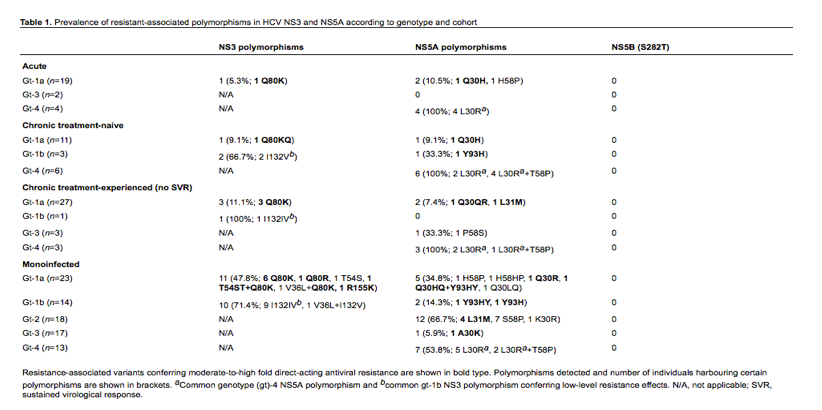
In this study, we investigated the prevalence of RAVs by population sequencing from three groups of HIV-HCV-coinfected patients: acute HCV infections (n=25), chronic treatment-naive patients (n=20) and chronic treatment-experienced (PEG-IFN/RBV) patients who did not achieve an SVR (n=34) and compared with the prevalence of RAVs in 85 chronic HCV-monoinfected patients. Genomic regions (sites of known RAVs) were amplified from HCV RNA using reverse transcriptase PCR followed by a nested PCR. Typically, amino acids 1-181 of HCV NS3 protease (gt1 only), amino acids 1-213 of domain I of NS5A (for gt-1a, 1b, 2, 3 and 4) and amino acids 219-347 of NS5B (pan-genotypic) were included. Purified PCR amplicons were sequenced using ABI PRISM 3730 genetic analyser (Applied Biosystems, Life Technologies Ltd, Paisley, UK) and consensus sequences aligned against HCV reference sequences.
Baseline RAVs were detected in all three cohorts of coinfected patients in NS3 and NS5A but baseline S282T, associated with resistance to sofosbuvir, was not detected in any of our cohorts (Table 1), possibly attributable to the low fitness of this mutation [10]. The Q80K polymorphism was the predominant NS3 variant for gt-1a conferring resistance to first- and second-generation PIs [11], and increased in frequency in acute versus chronic patients and from chronic treatment-naive to chronic treatment-failure patients: 5.3% versus 9.1% versus 11.1%, respectively. Conversely, the Q30H NS5A variant was only detected in a single acute patient, conferring 1477-fold DCV resistance [12] and variants at codon 30 did not differ markedly in HCV gt-1a between the chronic and acute cohorts. The NS5A Y93H variant was present in 33.3% compared with 7% reported by Plaza et al. [1] in our chronic treatment-naive gt-1b coinfected population, and was not detected in our HCV gt-1b treatment failure cohort. In contrast to Plaza et al. [1] we did not observe Y93H in any of our HCV gt-4-infected cohorts. However, similar to Plaza et al. [1], we observed in the majority of HCV gt-1b and gt-4 NS5 sequences, the M28L and L30R variants respectively, conferring minimal resistance effects to DCV. Unlike Plaza et al. [1], the L31M NS5A variant was not observed in our HCV gt-4a cohort but was detected in single HCV gt-1a treatment failure, reportedly conferring 341-fold DCV resistance effects [12]. Due to high viral load and increased rates of primary RAVs reported for HIV-HCV-coinfected patients versus HCV-monoinfected patients [13,14] we decided to compare the prevalence of DAA-resistant variants between these cohorts. We observed a significant increase in NS3 variants in HCV gt-1a monoinfected patients compared with acute and/or chronic HCV gt-1a coinfected patients (P<0.0001 Fisher's Exact test), including the R155K variant, conferring resistance to all licensed PIs [11,15]. We also observed an increase in NS5A variants in the monoinfected gt-1a cohort, though this was not statistically significant. Additionally, the L31M NS5A variant was detected in 22.2% HCV gt-2-monoinfected patients, conferring 140-fold DCV resistance, whilst the prevalence of NS3 and NS5A variants was minimal for HCV gt-3, with NS5A A30K variant observed in a single monoinfected patient.
In conclusion, natural polymorphisms conferring low-to-moderate NS3 and NS5A DAA resistance effects were detected across gts 1-4 in both HCV-monoinfected and acute and chronic HIV-HCV-coinfected patients, using population sequencing, hence probably under-estimating the presence of minority variants. The significance of this finding will only come to light as we start to put the use of new DAAs into clinical practice.
Low frequency of drug-resistant virus did not affect the therapeutic efficacy in daclatasvir plus asunaprevir therapy in patients with chronic HCV genotype 1 infection - see pdf attached above
|
|
| |
| |
|
|
|