 |
 |
 |
| |
EUROPEAN RAVS DATABASE: FREQUENCY AND CHARACTERISTICS OF RAVS IN TREATMENT-NAïVE AND DAA-EXPERIENCED PATIENTS
|
| |
| |
Reported by Jules Levin
EASL 2016 April 14-17 Barcelona
Simone Susser1, Julia Dietz* 1, Johannes Vermehren1, Kai-Henrik Peiffer1, Sandra Passmann1, Dany Perner1, Caterina Berkowski1, Peter Ferenci2, Maria Buti3, Beat Müllhaupt4, Bela Hunyadi5, Holger Hinrichsen6, Stefan Mauss7, Jorg Petersen8, Peter Buggisch8, Andreas Schober9, Gisela Felten10, Dietrich Hüppe10, Andreas Zipf11, Gaby Knecht12, Thomas Lutz12, Thomas Berg13, Stefan Zeuzem1, Christoph Sarrazin1
1Medizinische Klinik 1, Goethe-University Hospital, Frankfurt, Germany, 2Department of Internal Medicine III, Medical University of Vienna, Vienna, Austria, 3Hospital Universitario Valle Hebron and Ciberehd, Barcelona, Spain, 4Swiss Hepato-Pancreato-Biliary Center and Department of Gastroenterology and Hepatology, University Hospital Zürich, Zürich, Switzerland, 5Somogy County Kaposi Mór Teaching Hospital, Kaposvár, Hungary, 6Practice of Gastroenterology, Kiel, 7Practice of Gastroenterology, Düsseldorf, 8Institute for Interdisciplinary Medicine IFI, Hamburg, 9Practice of Hepatology, Gottingen, 10Practice of Hepatology, Herne, 11Practice of Gastroenterology, Mannheim, 12Infektiologikum, Frankfurt, 13Department of Gastroenterology and Rheumatology, University Hospital Leipzig, Leipzig, Germany
Program abstract
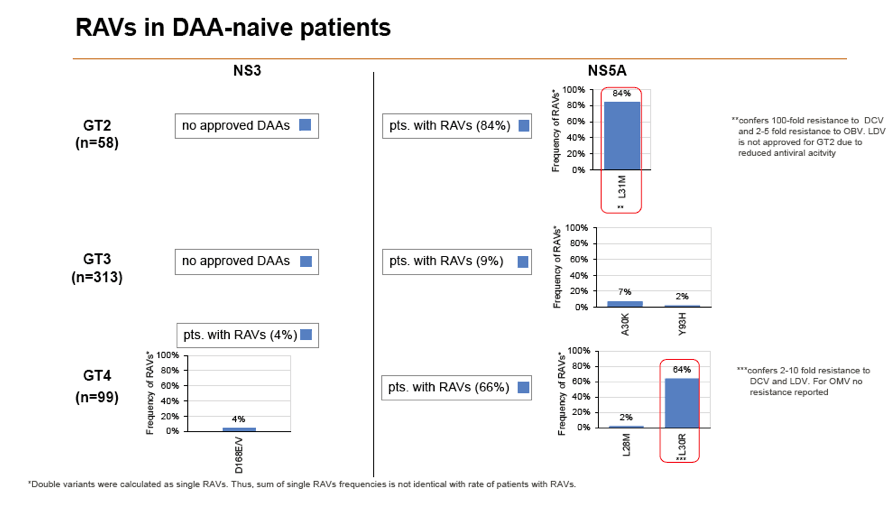
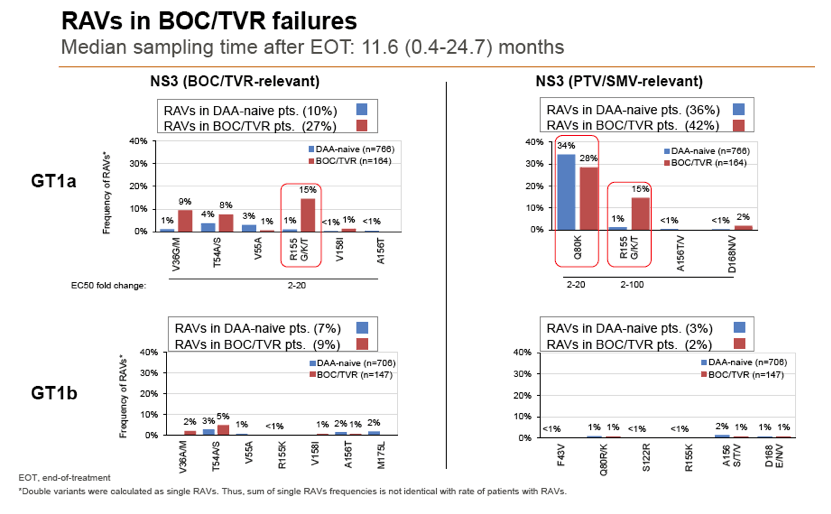
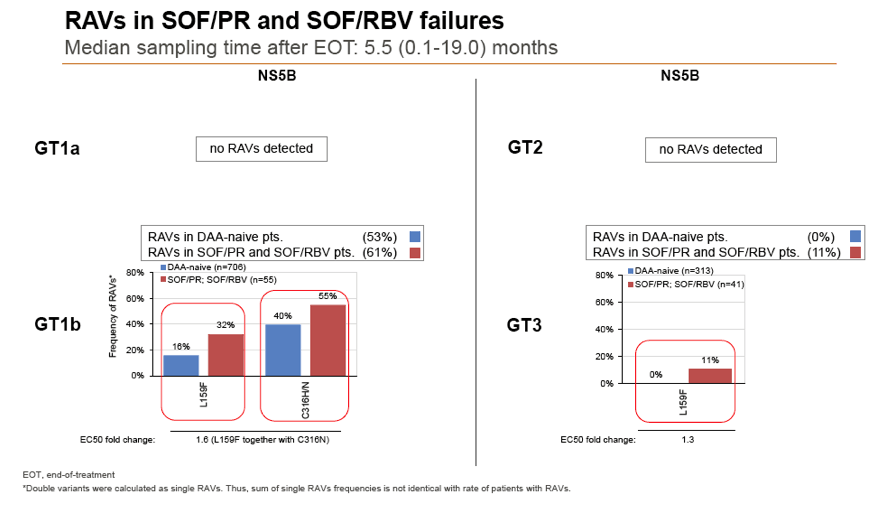
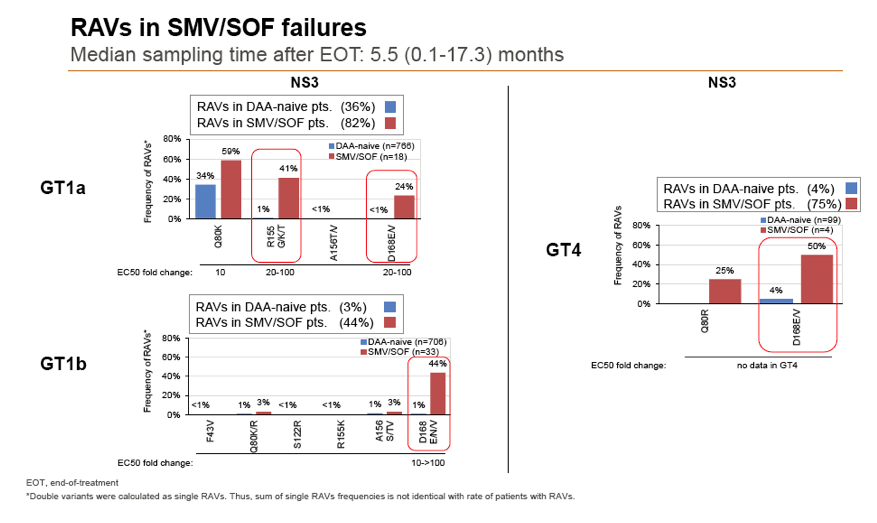
Background and Aims: Interferon-free combination therapies are the new standard for treatment of chronic hepatitis C virus (HCV) infection. The frequency of HCV RAVs (resistance associated variants) to direct antiviral agents (DAAs) varies between HCV genotypes, but pre-existing RAVs are often associated with virologic treatment failure. In this study frequencies of NS3, NS5A and NS5B RAVs were investigated in treatment-naïve and -experienced patients and consequences for DAA treatment options were evaluated.
Methods: Serum samples of 3305 European HCV infected patients were collected and population-based sequencing of HCV NS3, NS5A and NS5B genes was performed. RAVs were considered as relevant if they were associated with treatment failure or were shown to confer a >2-fold changed drug susceptibility in comparison to the reference strain. RAVs were analysed in NS3 (positions 36, 43, 54, 55, 56, 80,122, 155, 156,158,168, 170, 175), NS5A (24, 28, 30, 31, 58, 92, 93) and NS5B (159, 282, 321, 316, 368, 411, 414, 448, 553, 554, 556, 558, 559, 561).
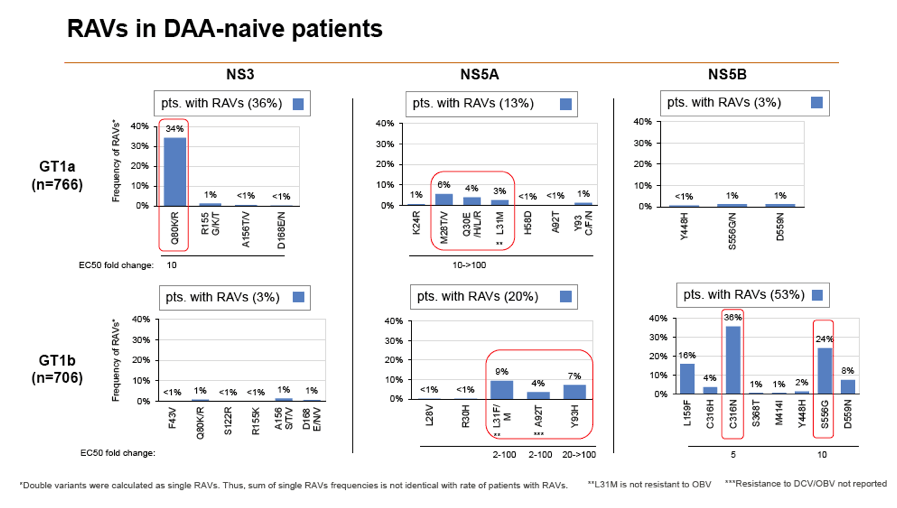
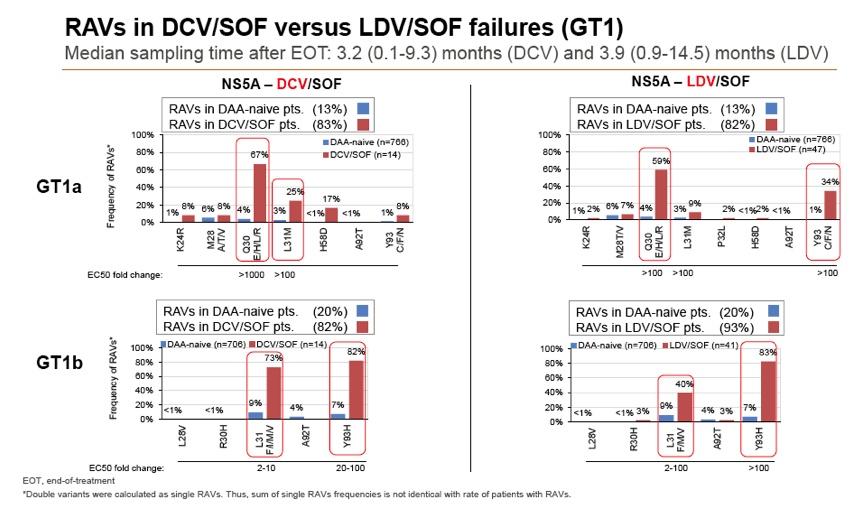
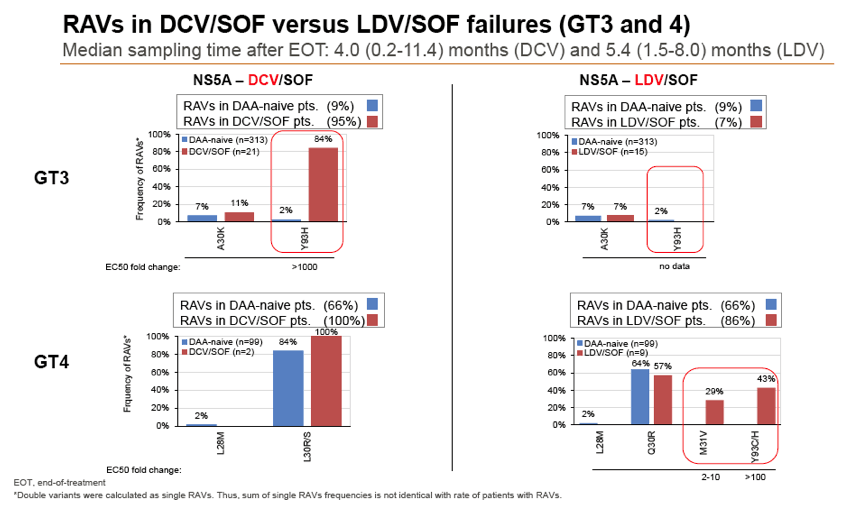
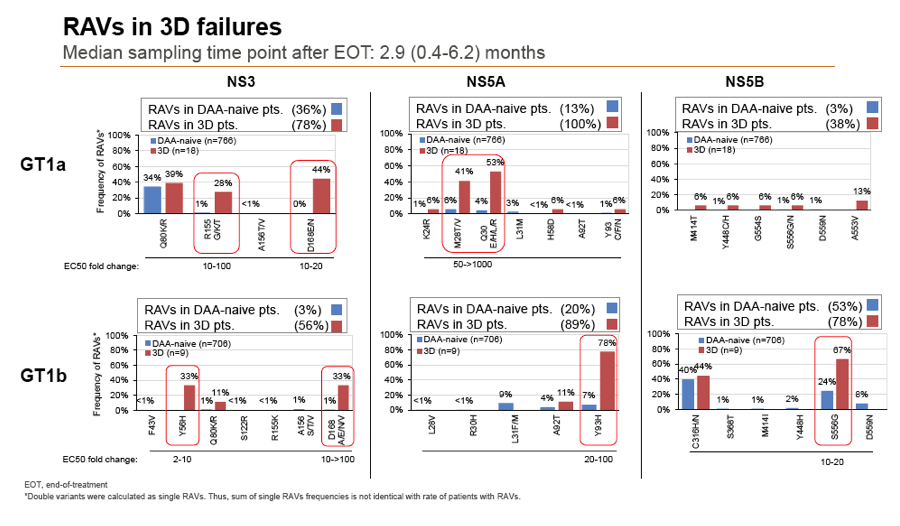
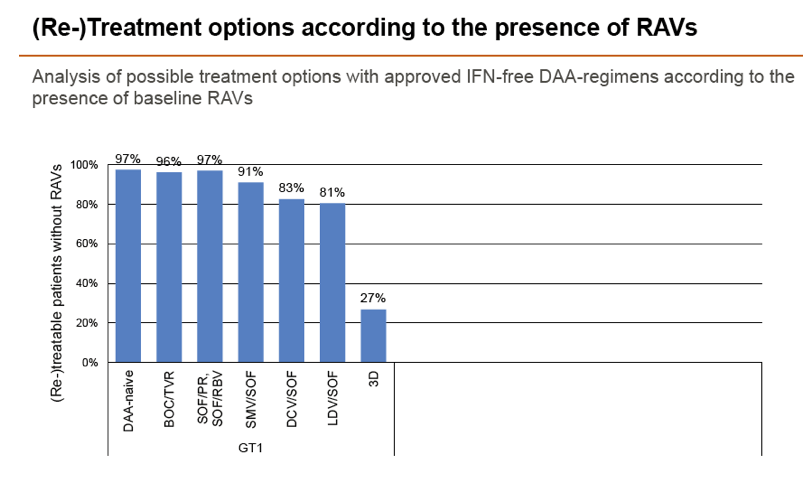
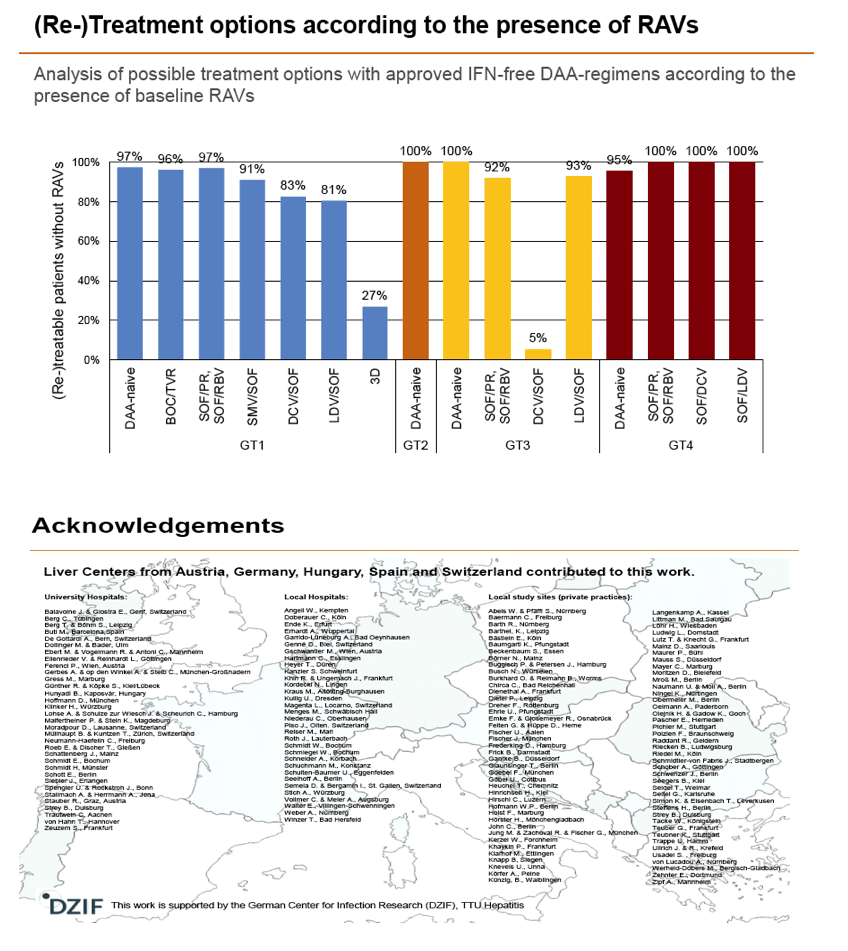
Results: Treatment-naïve and treatment-experienced patients infected with HCV genotype 1a (n = 1417), 1b (n = 1300), 1c-e (n = 5), 2 (n = 49), 3 (n = 389), 4 (n = 119), 5 (n = 7), 6 (n = 1), and 2k/1b (n = 18) were studied. Pre-existing RAVs could be observed in 38% of treatment-naïve patients. The proportion of selected RAVs in telaprevir, boceprevir and PEG/RBV pre-treated patients was 36%, 26%, and 34%, respectively. After failure to SOF/RBV ± PEG no RAVs could be detected. In patients treated with SOF in combination with SMV, DCV, or LDV a much higher incidence of RAVs was observed (64%, 84%, and 60%) only exceeded by PTV/OMB/DSV failure patients who all selected RAVs (100%). Re-/treatment without RAVs with currently approved regimens would be possible in 99% of the naïve patients and in 96% (TVR), 95% (BOC), 83% (SOF/RBV), 90% (SOF/PEG/RBV), 88% (SOF/SMV) 49% (SOF/DCV), 62% (SOF/LDV), 28% (PTV/OMB/DSV), and 98% (PEG/RBV) of pre-treated patients.
Conclusions: For treatment naïve patients RAVs against NS3, NS5A or NonNuc NS5B inhibitors are observed with moderate frequency (5-57%) and RAVs-free treatment options are available for almost all patients. However, in patients with failure to multiple DAA combination regimens, RAVs were found in 36-100% of patients which may impose restrictions on effective retreatment options with currently approved DAA regimens.
|
| |
|
 |
 |
|
|