| |
|
Difficult to treat Genotypes, sub-Saharan Africa, China or South-East Asia, migrants, Genotype Testing: EASL 2020 Recommendations
|
| |
| |
EASL recommendations on treatment of hepatitis C: Final update of the series
European Association for the Study of the Liver
Link to full....
EASL 2020 Recommendations on the treatment of Hepatitis C
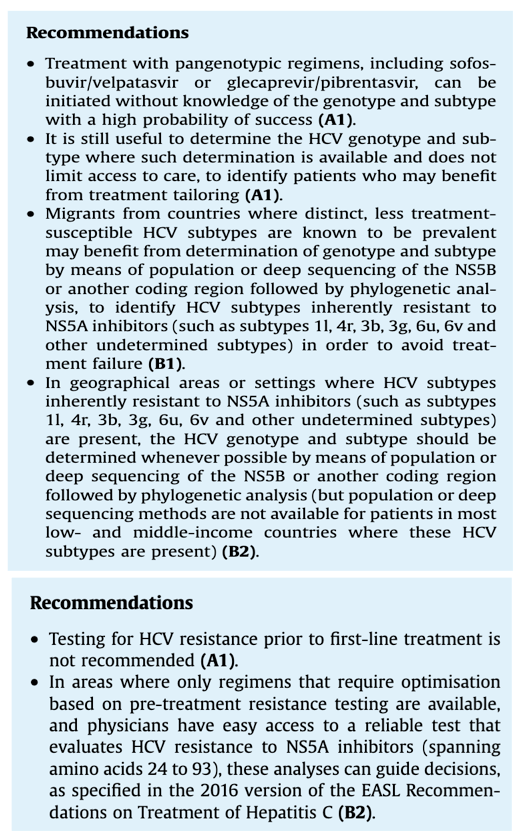
Migrants from countries where distinct, less treatment-susceptible HCV subtypes are known to be prevalent may benefit from determination of genotype and subtype by means of population or deep sequencing of the NS5B or another coding region followed by phylogenetic analysis, to identify HCV subtypes inherently resistant toNS5A inhibitors (such as subtypes 1l, 4r, 3b, 3g, 6u, 6v and other undetermined subtypes) in order to avoid treatment failure(B1)._ see tables below
In geographical areas or settings where HCV subtypes inherently resistant to NS5A inhibitors (such as subtypes1l, 4r, 3b, 3g, 6u, 6v and other undetermined subtypes) are present, the HCV genotype and subtype should be determined whenever possible by means of population or deep sequencing of the NS5B or another coding region followed by phylogenetic analysis (but population or deep sequencing methods are not available for patients in most low- and middle-income countries where these HCV subtypes are present) (B2). See tables below
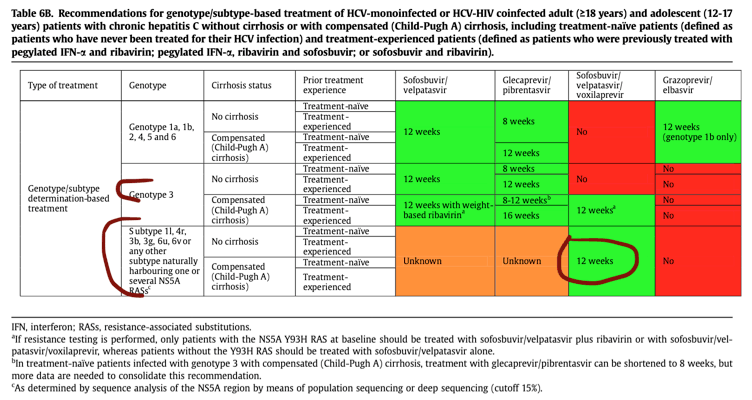
In settings where sequence analysis of the NS5A region by means of population sequencing or deep sequencing (cut-off15%) is available and affordable, it should be performed in patients born in sub-Saharan Africa, China or South-East Asia in order to: (i) identify infrequent HCV subtypes not detected by the line probe assay (defined as genotype 1 non-1a/1b, genotype2 non-2a/2b, genotype 3 non-3a, genotype 4 non-4a/4d, an dsubtypes of genotypes 5 to 8) by means of phylogenetic analysis of the sequences generated, and (ii) characterize the NS5A RAS profile to identify patients harbouring viruses inherently resistant to NS5A inhibitors. In the absence of clinical trial or real-world data, patients infected with subtypes 1l, 4r, 3b, 3g, 6u and 6v and patients infected with other infrequent subtypes harbouring >-1 RAS(s) known to confer resistance to NS5A inhibitors (see below) should be treated firstline with the fixed combination of sofosbuvir, velpatasvir and voxilaprevir, pending data with dual pangenotypic regimens. The following genotype/subtype-dependent treatment recommendations are summarised in Table 6B: see table 6 below
In settings where sequence analysis of the NS5A region by means of population or deep sequencing is available and affordable, patients infected with subtypes 1l, 4r, 3b, 3g, 6u and 6v and patients infected with other infrequent subtypes harbouring>-1 RAS(s) known to confer resistance to NS5A inhibitors should be considered for treatment with the fixed-dose combination of sofosbuvir, velpatasvir and voxilaprevir for 12 weeks, pending data with dual pangenotypic regimens(B2). See table 6 below
HCV genotype determination
Pan-genotypic HCV drug regimens, including sofosbuvir/velpatasvir and glecaprevir/pibrentasvir, can be used to treat in-dividuals without identifying their HCV genotype and subtype, simplifying therapy.
Nevertheless, identifying certain genotypes before starting first-line therapy remains useful and may be required where drug procurement or pricing dictates genotype-specific treatment, or to optimise treatment regimens. Genotyping/subtyping should be performed with an assay that accurately discriminates subtype 1a from 1b, i.e. an assay using the sequence of the 50 untranslated region plus a portion of another genomic region, generally the core-coding or the NS5B-coding regions.104 The most widely used, CE-IVD-marked method is based on reverse hybridisation with the second-generation line probe assay.105 A
commercial CE-IVD-marked assay based on deep sequencing is also available.106,107
Recently, distinct subtypes of genotypes 1 to 8 that are in frequent in Europe, North America, Japan and Australia (defined as genotype 1 non-1a/1b, genotype 2 non-2a/2b, genotype 3 non-3a, genotype 4 non-4a/4d, and subtypes of geno-types 5 to 8) have been shown to be highly prevalent in certainregions of Africa and Asia and in migrants from these regions. 108–111 Some (for instance genotypes 1l, 4r, 3b, 3g, 6u, 6va mong others) harbour natural polymorphisms that confer inherent resistance to NS5A inhibitors, resulting in unacceptably frequent virological failures in both the resident populations as well as in migrants from these regions.108,110–115 Thus, HCV genotype and subtype should ideally be determined before treatment in regions where these HCV subtypes are present insubstantial proportions, or in migrants from these regions, tooptimise treatment regimens. Reverse hybridisation with the line probe assay accurately identifies only genotypes 1 to 6 and subtypes 1a and 1b, but misclassifes most of these infrequent, less treatment-susceptible subtypes.105 Their accurate determination requires sequence analysis of the NS5B or another coding region of the HCV genome followed by phylogenetic analysis. A commercial CE-IVD-marked assay based on deep sequencing can be used for this purpose, but it requires specific equipment and skills.106,107 If this assay is not available and/or not affordable, only in-house population sequencing (Sanger sequencing) or deep sequencing technologies can be used. These technologies are not available in low- and middle-income settings where these subtypes are prevalent. Virological studies are required in countries in Africa, Asia and South America to determine the epidemiology, distribution and prevalence of HCV subtypes inherently resistant to NS5A inhibitors and thus to optimise treatment decisions without the need for individual HCV genotype and subtype determination.1
| |
| |
| |
|
|
|