 |
 |
 |
| |
HIV-1 DNA GENOTYPING IS OFTEN VARIABLE IN REPEAT TESTING FROM SINGLE BLOOD DRAWS
|
| |
| |
CROI 2021 March 6-10 Reported by Jules Levin
Michelle L . D'Antoni1, Kristen Andreatta1, Rima K. Acosta1, Hui Liu1, Yongwu Shao1, Kirsten L. White1
1Gilead Sciences, Inc, Foster City, CA, USA
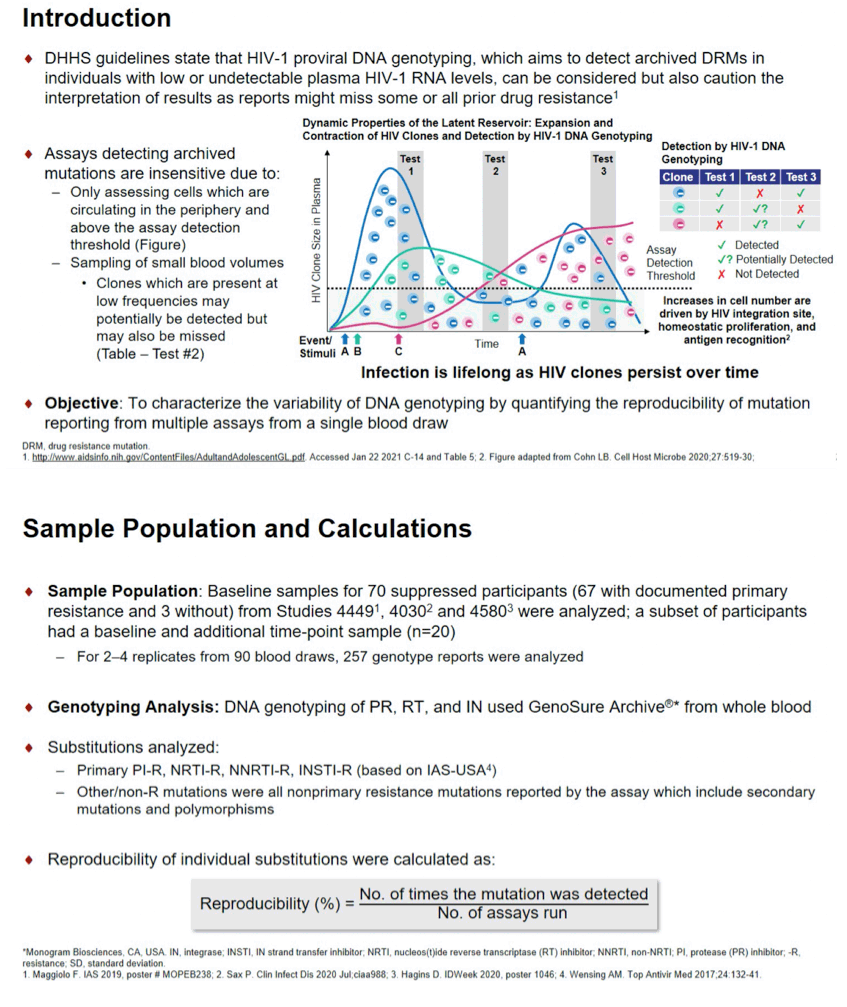
Background: HIV-1 DNA genotyping assesses archived drug resistance mutations (DRMs) in individuals with low plasma HIV RNA; however, assays detecting these mutations are insensitive. Here we seek to characterize the variability of DNA genotyping by quantifying the reproducibility of mutation reporting from multiple assays from a single blood draw.
Methods: DNA genotyping of protease (PR), reverse transcriptase (RT) and integrase (IN) used GenoSure Archive® (Monogram Biosciences, CA, USA) from whole blood from suppressed participants with documented resistance from 3 clinical trials (NCT03631732; NCT03110380; NCT03405935). Multiple tests (2-4) were run from each whole blood sample. Reproducibility of primary PR inhibitor (PI)-resistance (-R), nucleos(t)ide RT inhibitor (NRTI)-R, non-NRTI (NNRTI)-
R, IN strand transfer inhibitor (InSTI)–R, and other/non-R mutations were calculated as the number (#) of times the mutation was detected/# of assays run (%). Means ±standard deviations (SD) were reported, and comparisons used Wilcoxon Rank Sum tests. A zero-truncated binomial model was used to estimate the probability of mutation detection.
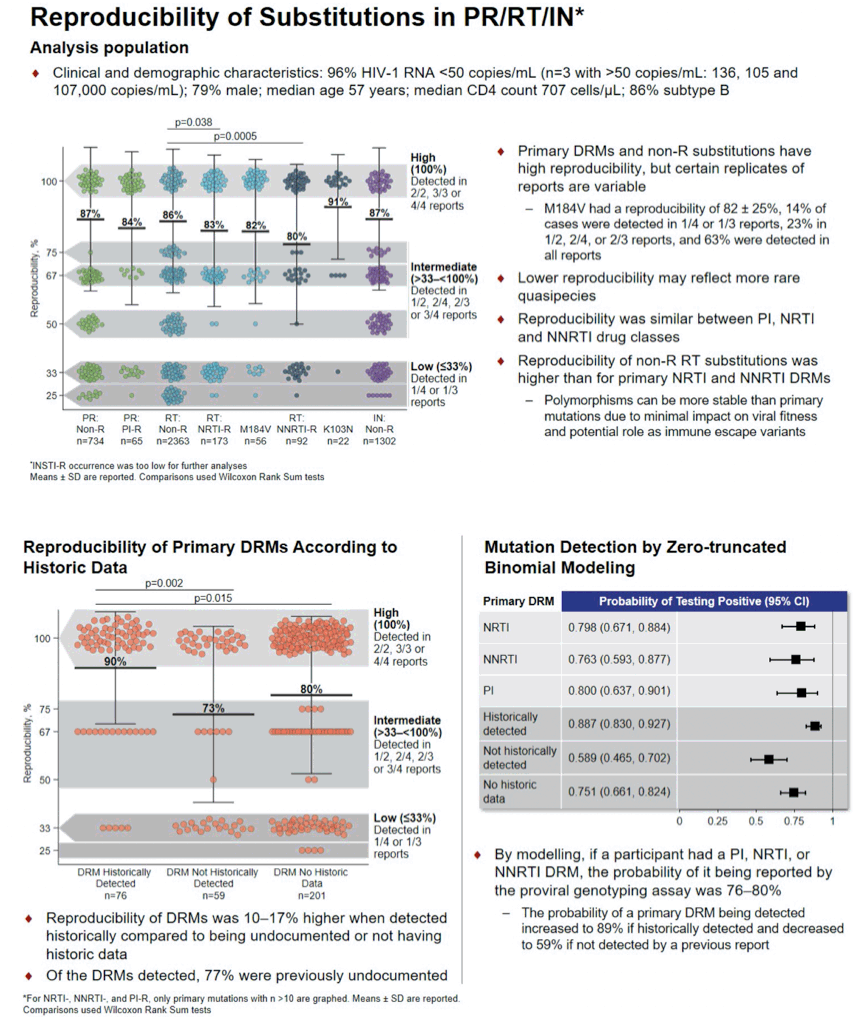
Results: For 90 blood draws from 70 participants (79% male; age 56 y, 714 CD4 count, 86% subtype B), 257 genotype reports were analyzed. Overall, reproducibility was similar for all PR, RT and IN mutations (86±25%, 86±25% and 87±25%, respectively). A total of 15 PI, 18 NRTI and 19 NNRTI primary DRMs were detected in 21, 43, and 31 participants, respectively, with reproducibility of 84±27%, 83±26%, 78±28% (Fig). InSTI-R occurrence was too low (n=2) for further analyses. The NRTI DRM M184V had a reproducibility of 82±25%, with 14% of cases being detected in 1/4 or 1/3 reports, 23% in 1/2, 2/4, or 2/3 reports, and 63% being detected in all reports. Reproducibility did not differ among drug classes. Reproducibility of polymorphisms and other non-R RT mutations was significantly higher than for primary NRTI and NNRTI DRMs (p<0.05).
Reproducibility of primary DRMs was 10-16% higher when detected by historical genotype compared to not being reported or not having data (p<0.05). By modelling, if a person had a PI, NRTI, or NNRTI DRM, the probability of it being reported by the assay was 76-80%.
Conclusion: Mean reproducibility of ∼80% with standard deviations of ∼25% indicate that detection of mutations is variable. DNA genotyping may aid clinicians when switching HIV regimens, but these data reinforce the need to interpret tests with caution, as not all mutations may be reported.

|
| |
|
 |
 |
|
|