 |
 |
 |
| |
ULTRASENSITIVE HIV-1 DRUG-RESISTANCE ANALYSIS IN THE DISCOVER PrEP TRIAL
|
| |
| |
CROI 2021 March 6-10 Reported by jules Levin
Stephanie Cox1, Urvi Parikh2, Amy Heaps2, Jay Goetz2, John W. Mellors2, Moupali Das1, Christian Callebaut1 1Gilead Sciences, Inc, Foster City, CA, USA, 2University of Pittsburgh, Pittsburgh, PA, USA
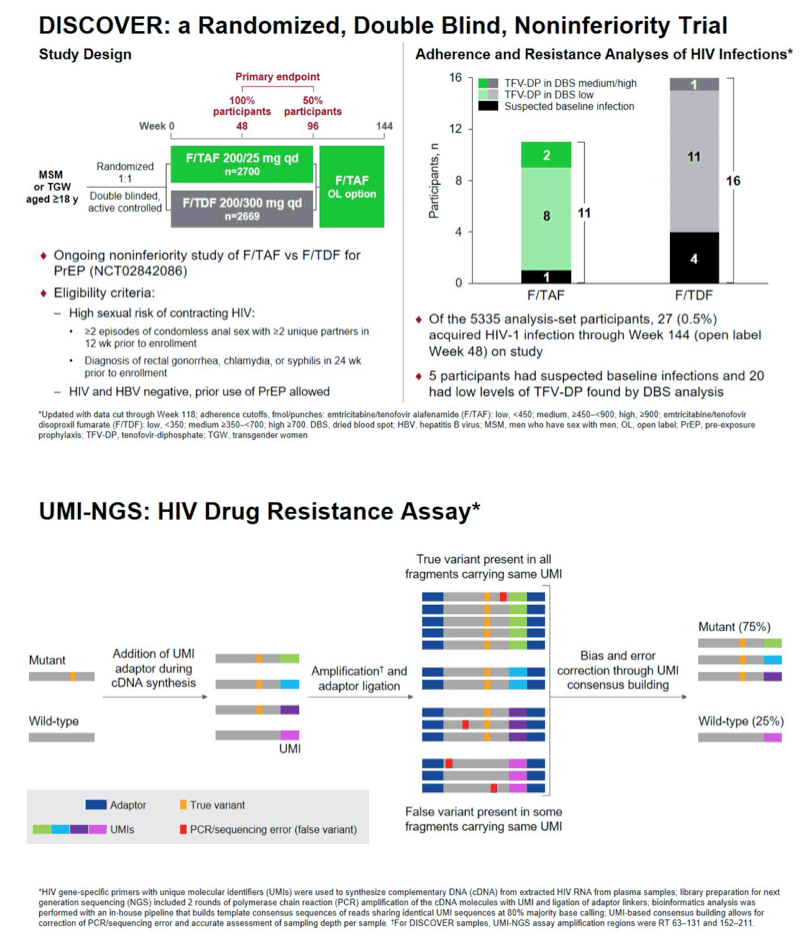
Background: The DISCOVER study is an ongoing randomized, double blind study of pre-exposure prophylaxis (PrEP) using daily FTC/TAF (F/TAF; Descovy; DVY) or FTC/TDF (F/TDF; Truvada; TVD) in men or transgender women who have sex with men. Of the 5,335 randomized participants evaluated for HIV-1 infection, 27 participants (0.5%) became infected with HIV-1 through 144 weeks on study. Participants who acquired HIV were evaluated with population, standard next generation sequencing (NGS), and ultrasensitive sequencing and the overall resistance data and analysis are presented here.
Methods: Plasma samples from participants who became infected with HIV-1 and had a viral load of > 400 copies/mL were tested with the GenoSureTM MG assay (Monogram) to analyze the protease (PR) and reverse transcriptase (RT) genes for resistance associated mutations (RAMs) (at ≥15-20% of the viral population). Additional analysis of the PR-RT and integrase (IN) genes on all available plasma samples was performed by standard NGS (SeqIT) to evaluate resistance mutations (at ≥2% of the viral population). Ultrasensitive resistance testing was done using unique molecular identifiers for amplification of viral variants (at ≥1% of the viral population) followed by NGS (UMI-NGS), to analyze RT codons 63-131 and 152-211 (University of Pittsburgh).
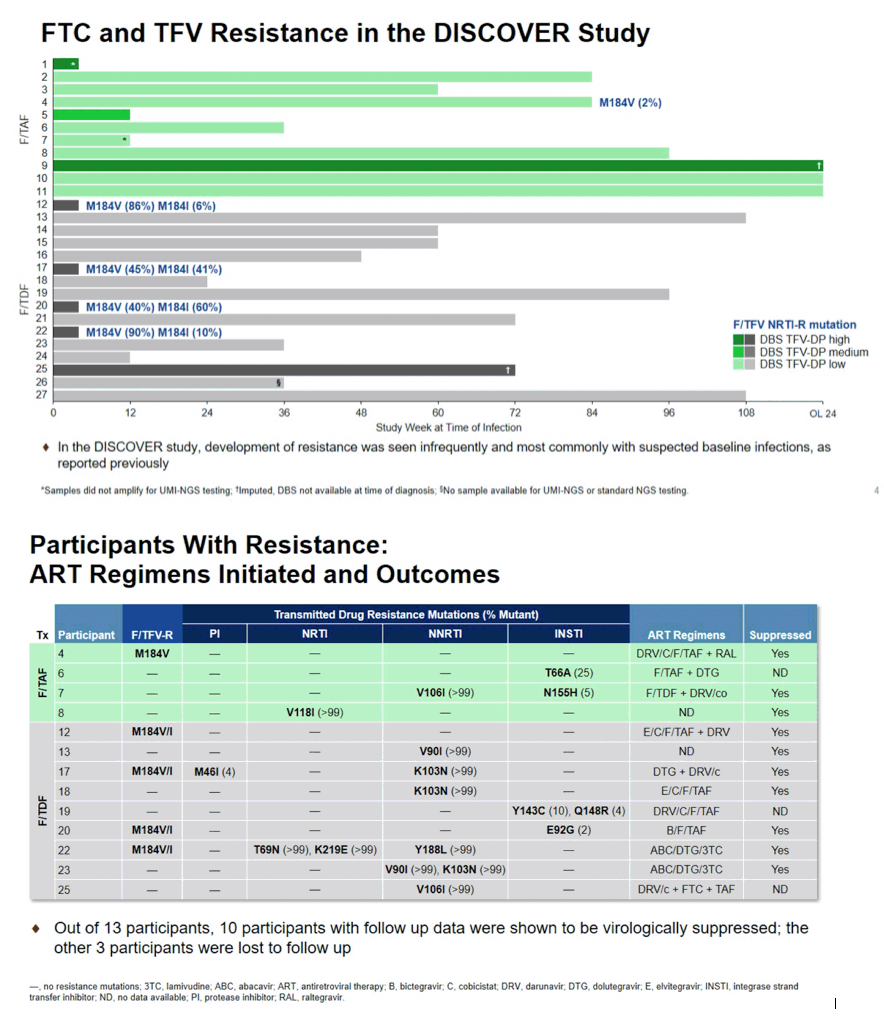
Results: By population sequencing, 4/20 participants infected with HIV had M184V, all in the F/TDF group and all with suspected baseline infection; 2 of these 4 also had M184I present. By standard NGS and UMI-NGS, 26/27 HIV participants infected with HIV had samples available and 25/27 were successfully analyzed. For the 4 participants on F/TDF with M184V, each had M184I also detected. By UMI-NGS, 1 participant on F/TAF had the M184V mutation present at 2%. Ten participants had additional mutations conferring resistance to non-study drugs including InSTI RAMs T66A, E92G, Y143C, Q148R, N155H; PI RAMs M46I; NNRTI RAMs V90I, V106I, K103N, Y188L, which were presumed to be transmitted.
Conclusion: Using population sequencing and standard NGS, M184V was detected in 4 participants, all in the F/TDF arm. With ultrasensitive UMI-NGS testing, similar results were observed in the F/TDF arm, with the addition of 1 participant with M184V in the F/TAF arm. In addition, analysis by standard NGS of the PR, RT and IN genes found notable transmitted drug resistance to non-study drugs. Overall, resistance to study drugs in the DISCOVER study was infrequently seen and primarily with suspected baseline infections.

|
| |
|
 |
 |
|
|